Content
Experiment
COLTRIMS (Cold Target Recoil Ion Momentum Spectroscopy)
Data
on 2019-06-25 runs 91, 92, 93, 100 requested to restore from tape
/reg/d/psdm/AMO/amox27716/xtc/
exp=amox27716:run=100
event_keys -d exp=amox27716:run=100
Scripts and files
For exp=amox27716:run=100 in LCLS1 environment
- hexanode/examples/ex-quad-00-data.py - simple psana access to data and draws waveforms overlayed on the same plot
- hexanode/examples/ex-quad-01-cfd.py <Acqiris-chanel-number> - test of CFD (Constant Fraction Discriminator) parameters
- hexanode/examples/ex-quad-02-amox27716-0100-acqiris.py - advanced test of CFD with event loop and graphics
- hexanode/examples/ex-quad-07-proc-data-save-h5.py - processes waveforms in dataset and saves results for peaks in file like amox27716-r0100-e060000-single-node.h5
- hexanode/examples/ex-quad-09-sort-graph-data.py
- uses processed data from amox27716-r0100-e060000-single-node.h5 or raw (slow) exp=amox27716:run=100 and calibration files- /reg/d/psdm/amo/amox27716/calib/Acqiris::CalibV1/AmoEndstation.0:Acqiris.1/hex_config/0-end.dat - control anf config file
copy of /reg/d/psdm/xpp/xpptut15/calib/Acqiris::CalibV1/AmoETOF.0:Acqiris.0/hex_config/0-end.data - /reg/d/psdm/amo/amox27716/calib/Acqiris::CalibV1/AmoEndstation.0:Acqiris.1/hex_table/100-end.dat - table with results of the detector calibration
- /reg/d/psdm/amo/amox27716/calib/Acqiris::CalibV1/AmoEndstation.0:Acqiris.1/hex_config/0-end.dat - control anf config file
- hexanode/examples/ex-quad-10-sort-data.py - TBD
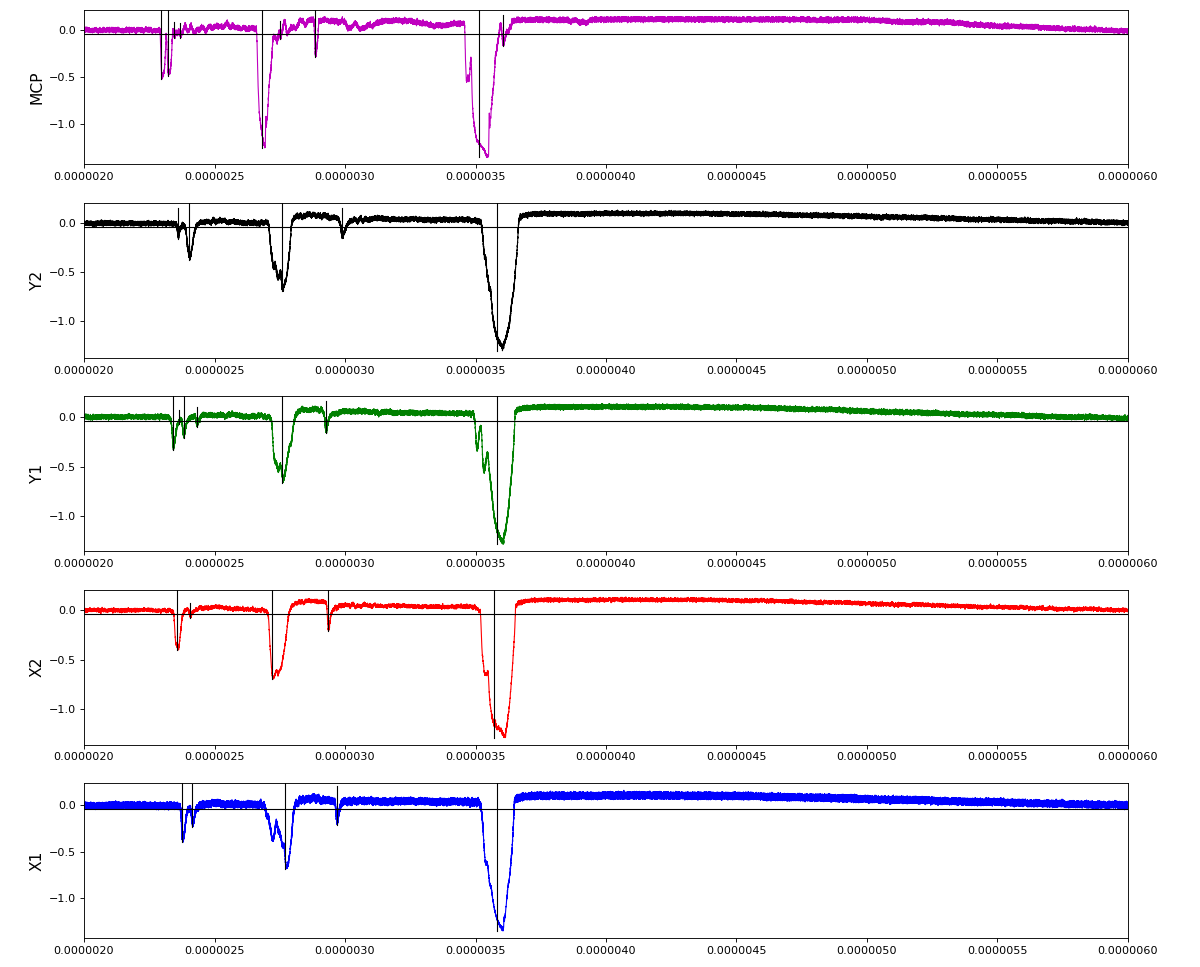
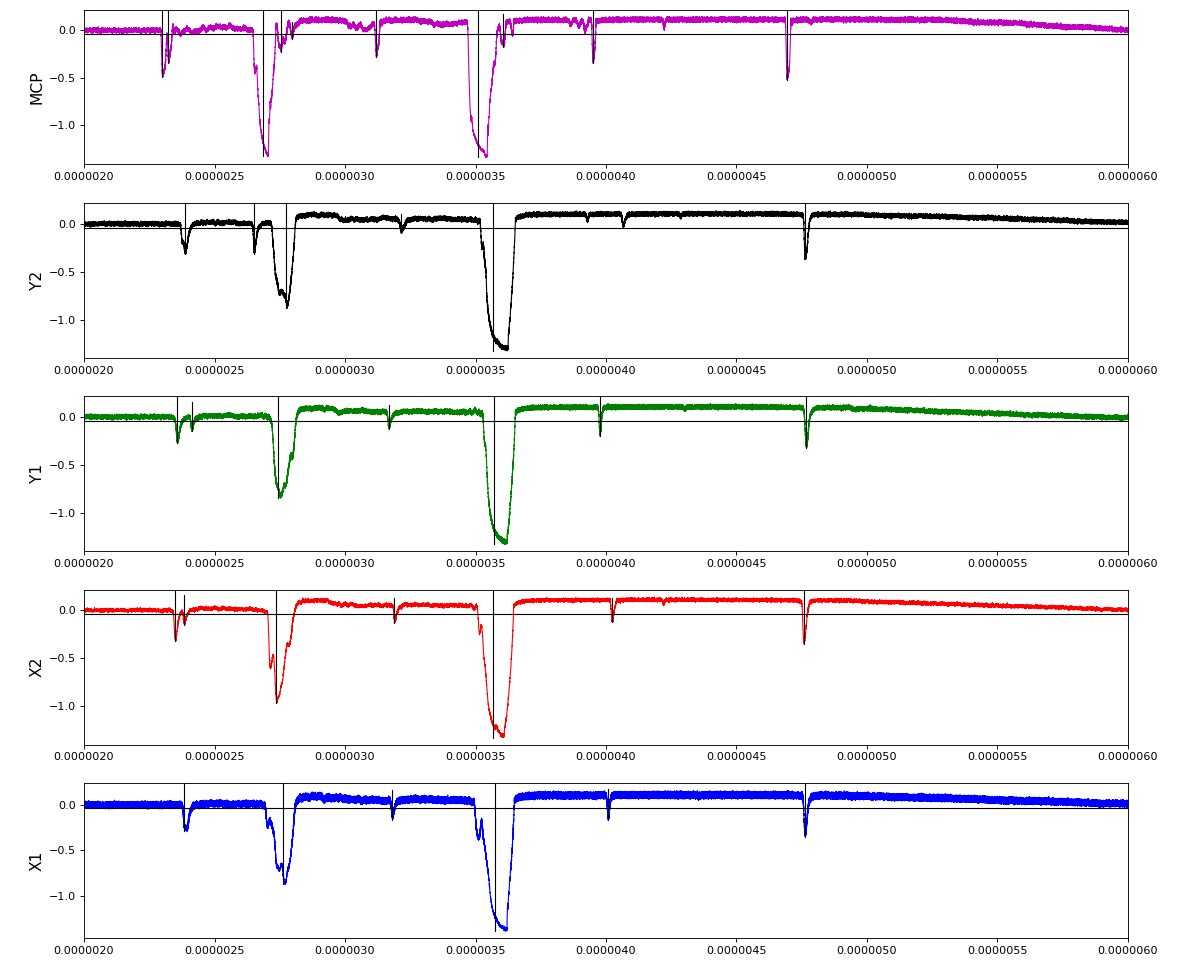
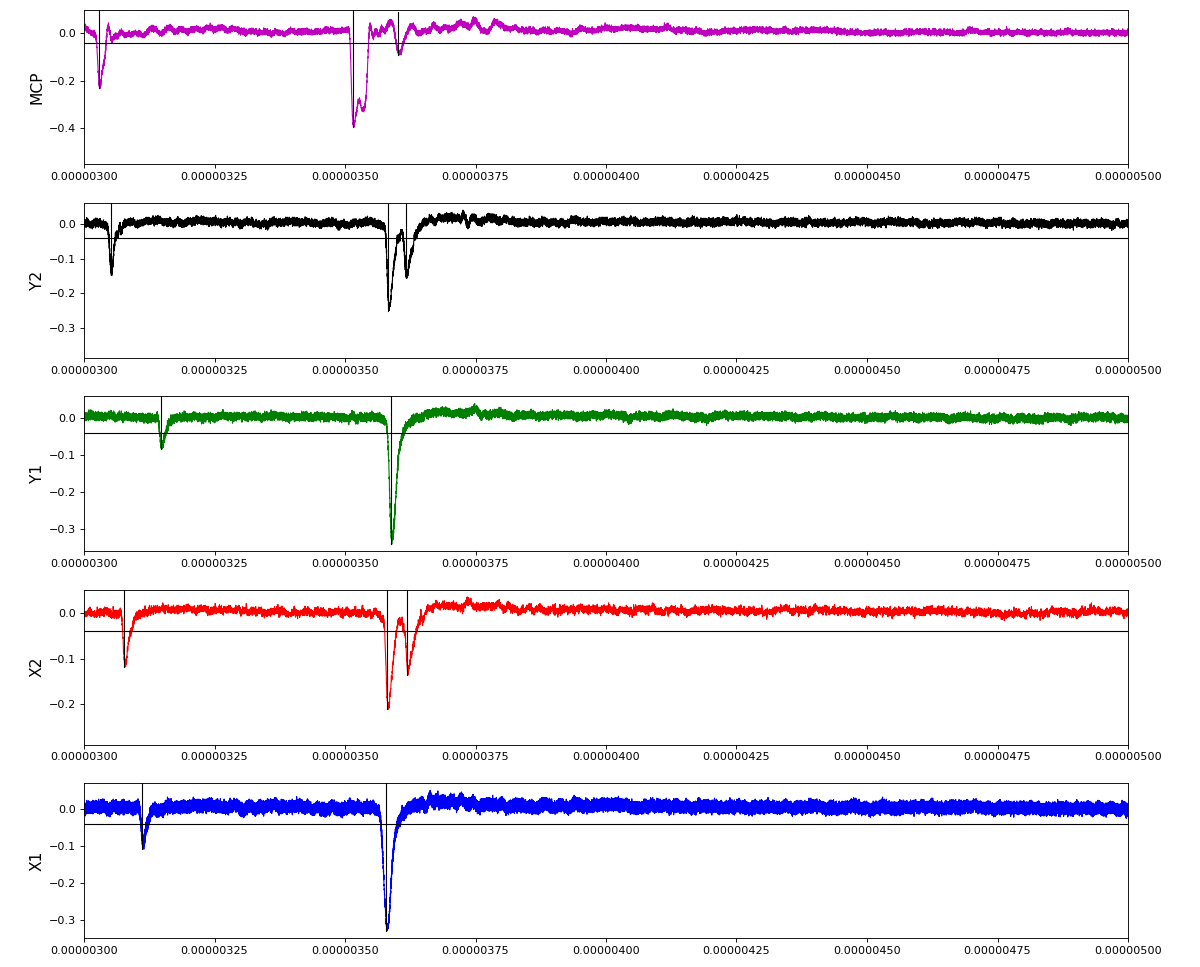
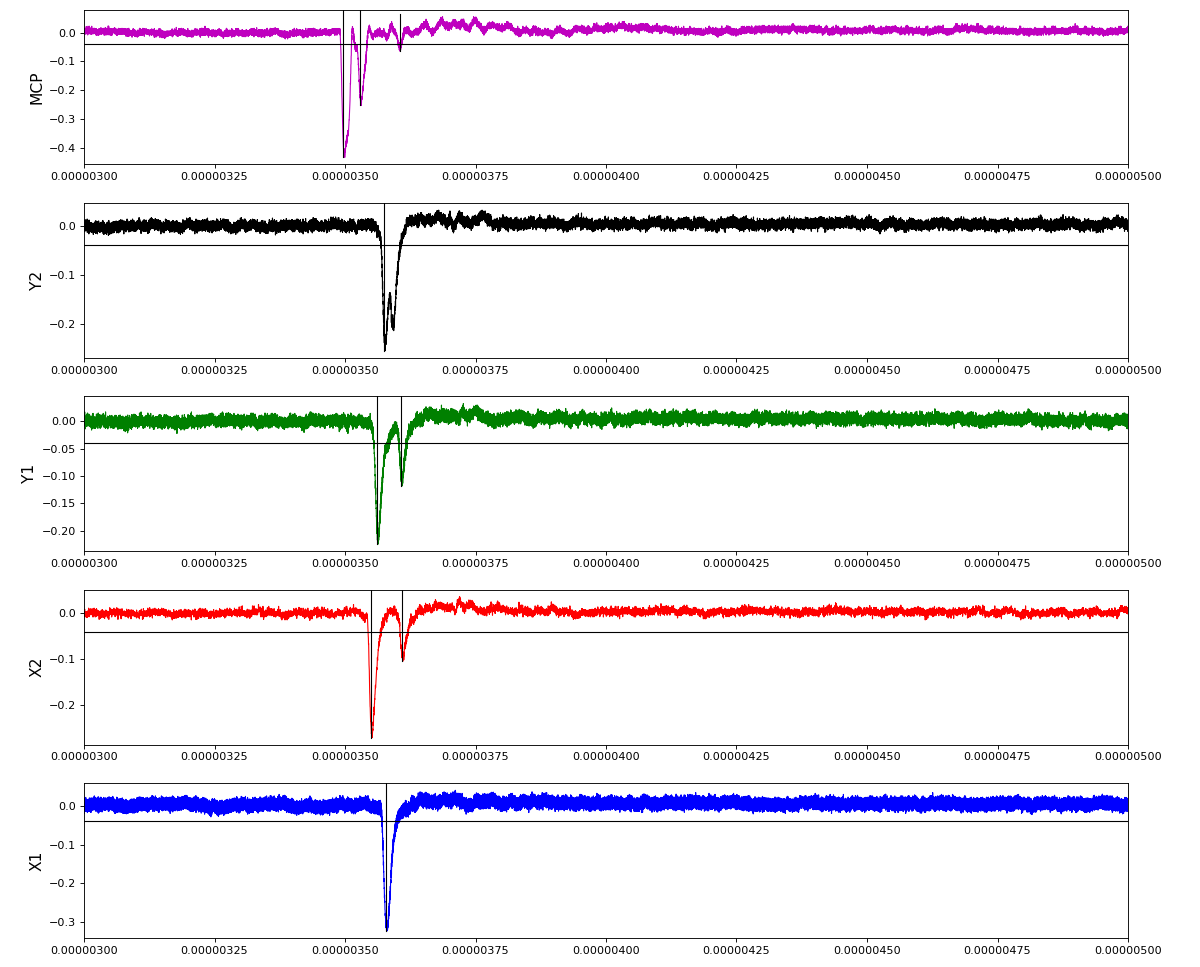
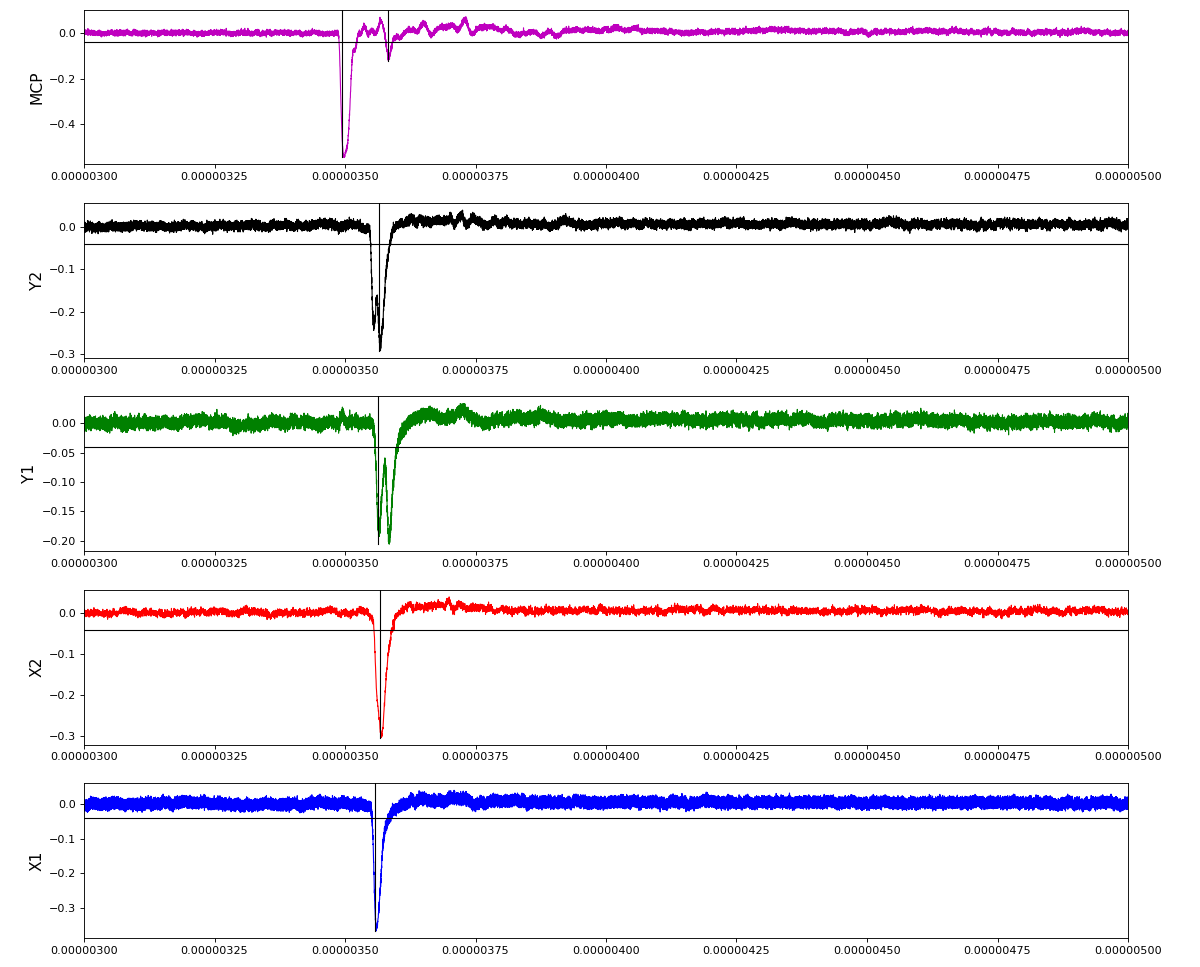
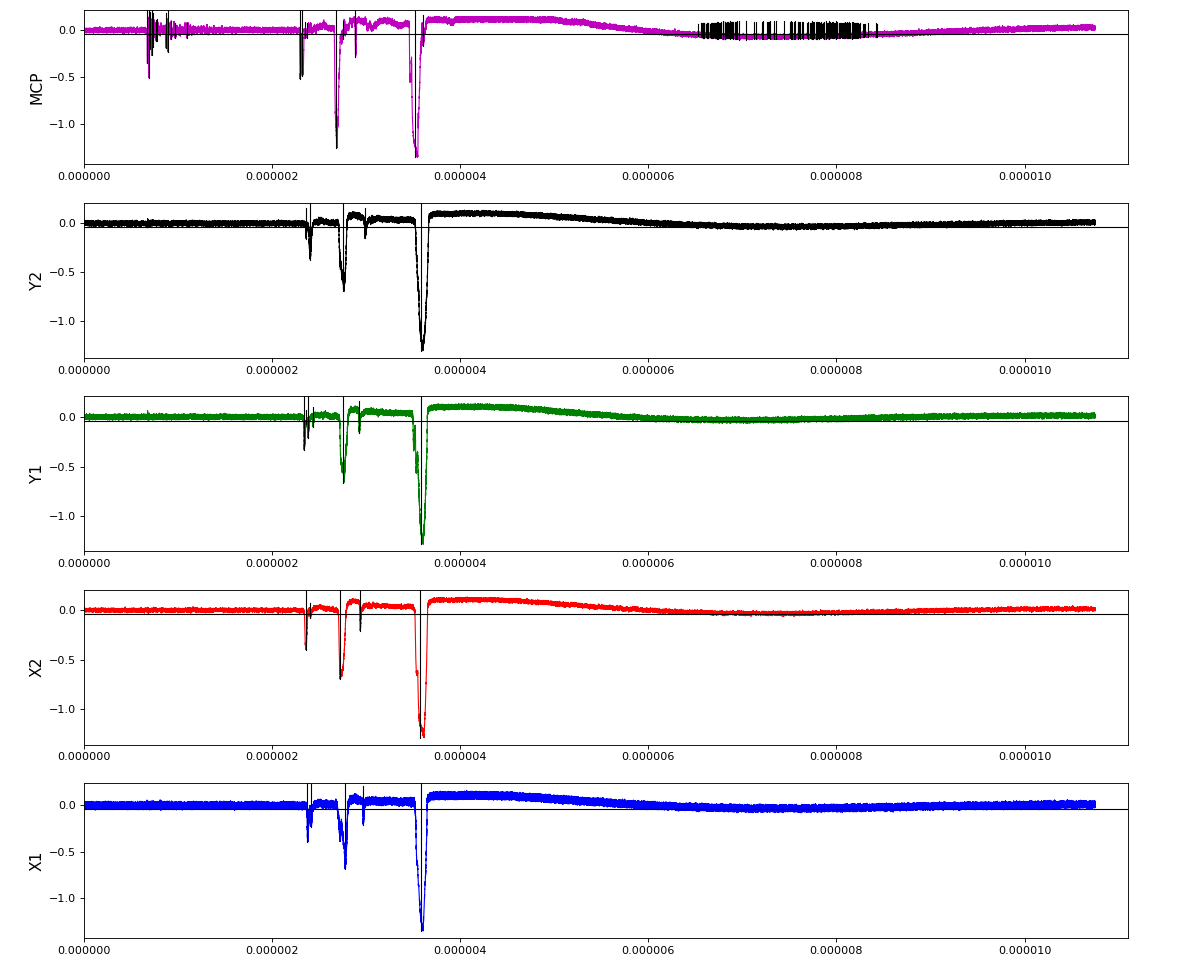
Waveforms
hex/examples/ex_acqiris_quad_amox27716-0100.py
Association of channels of AmoEndstation.0:Acqiris.1
ch = (2,3,4,5,6) - all other channels of Acqiris.1 and 2 are empty or noisy
ylab = ('X1', 'X2', 'Y1', 'Y2', 'MCP')
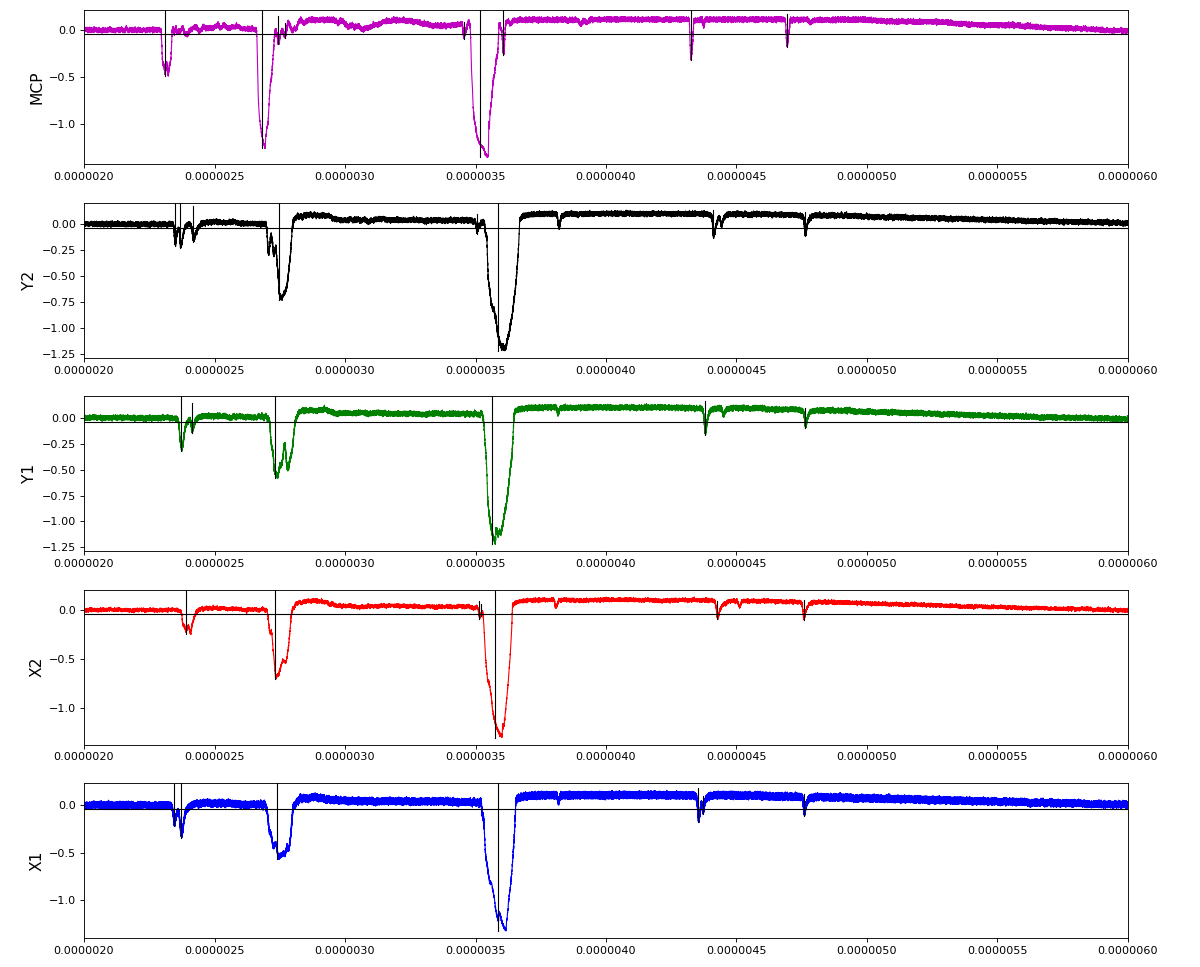
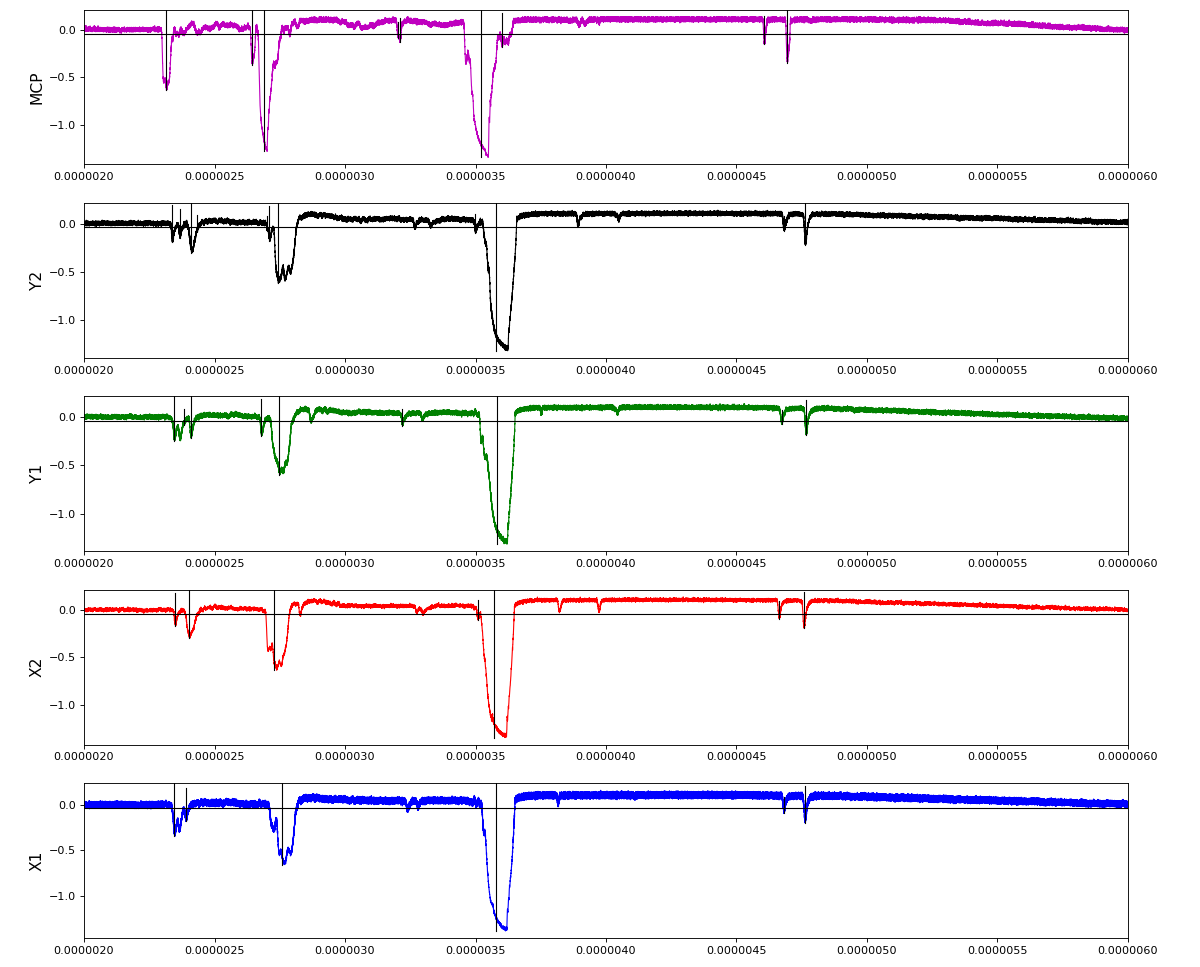
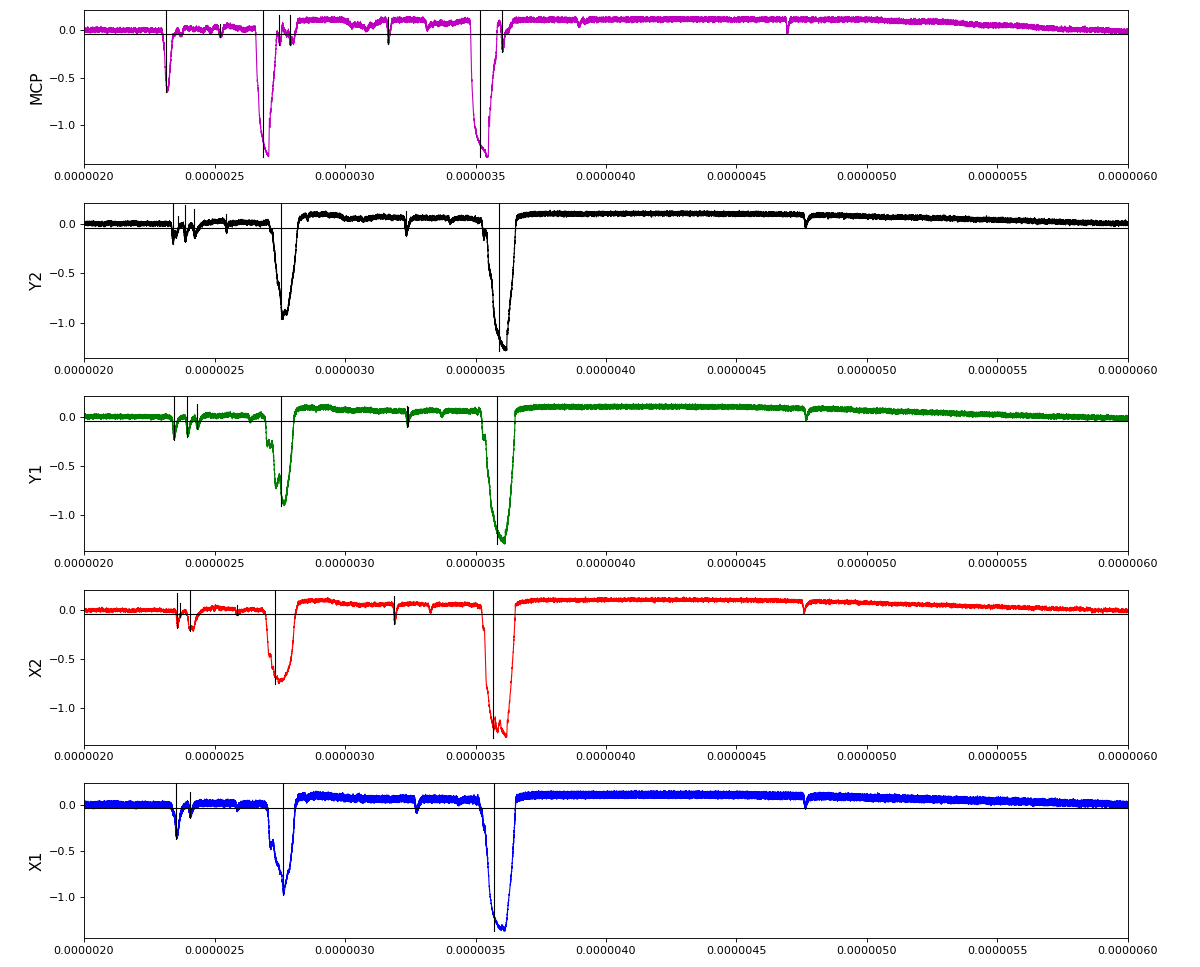
Other run amox27716 run 91 with lower hit density
Data processing
- hexanode/examples/ex-quad-02-amox27716-0100-acqiris.py - play with waveform parameters and the part of waveform WITHOUT NOISE!
- hexanode/examples/ex-quad-07-proc-data-save-h5.py - set w-f selection parameters data, detector channels, run over data and create hdf5 file.
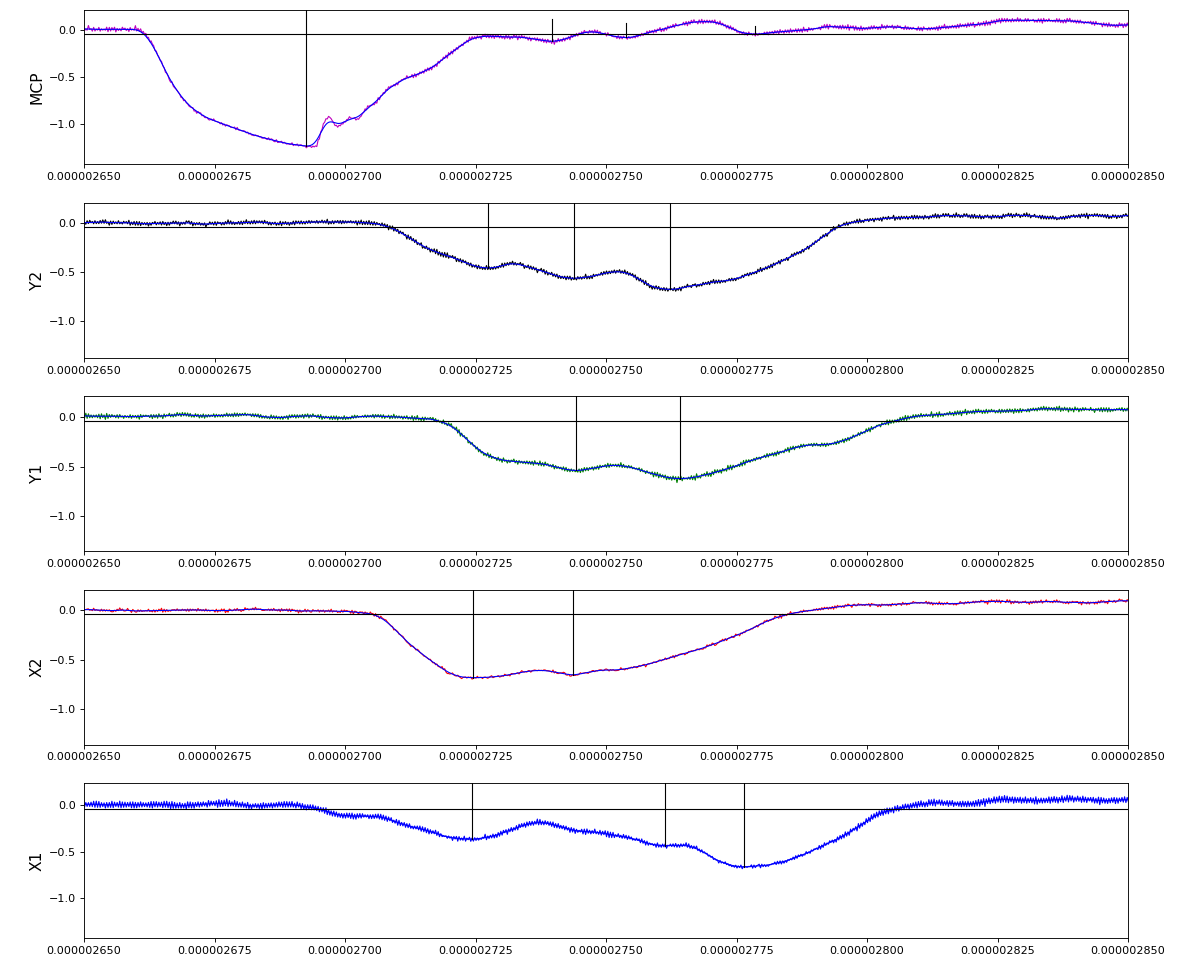
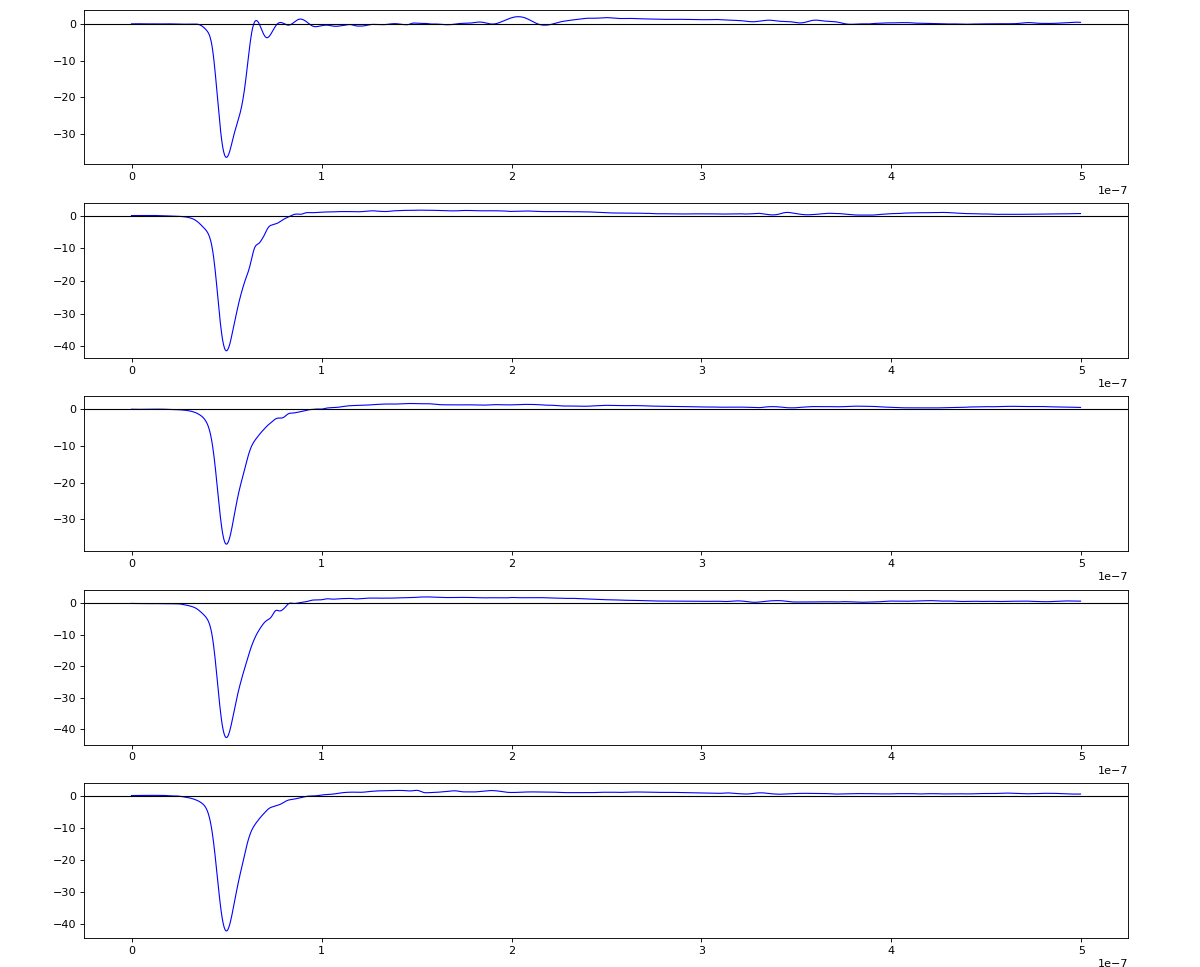
Total waveform with problematic peak reconstruction in MCP channel:
Test of local extreme peak-finder
Version of peak finder searching for local extremes after filtering
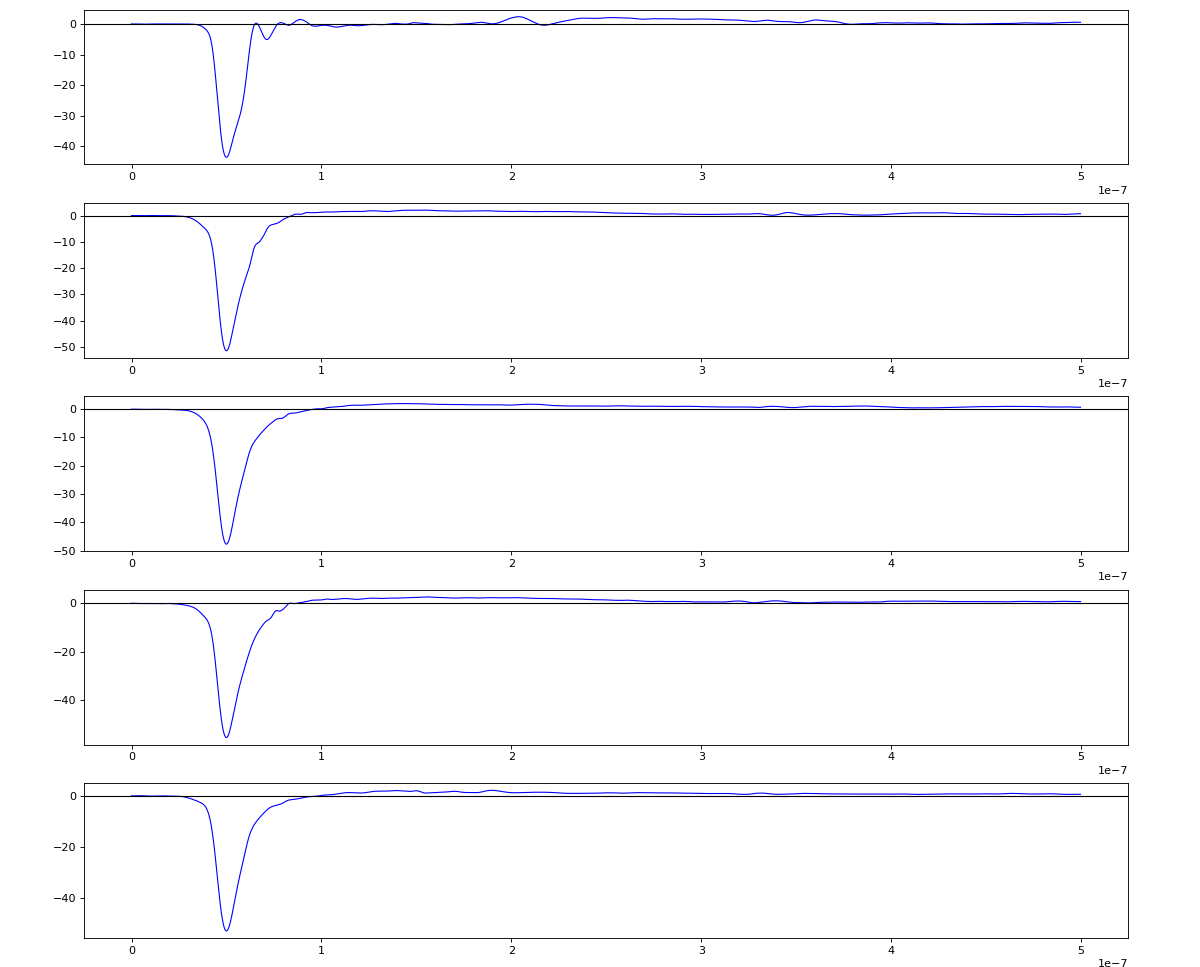
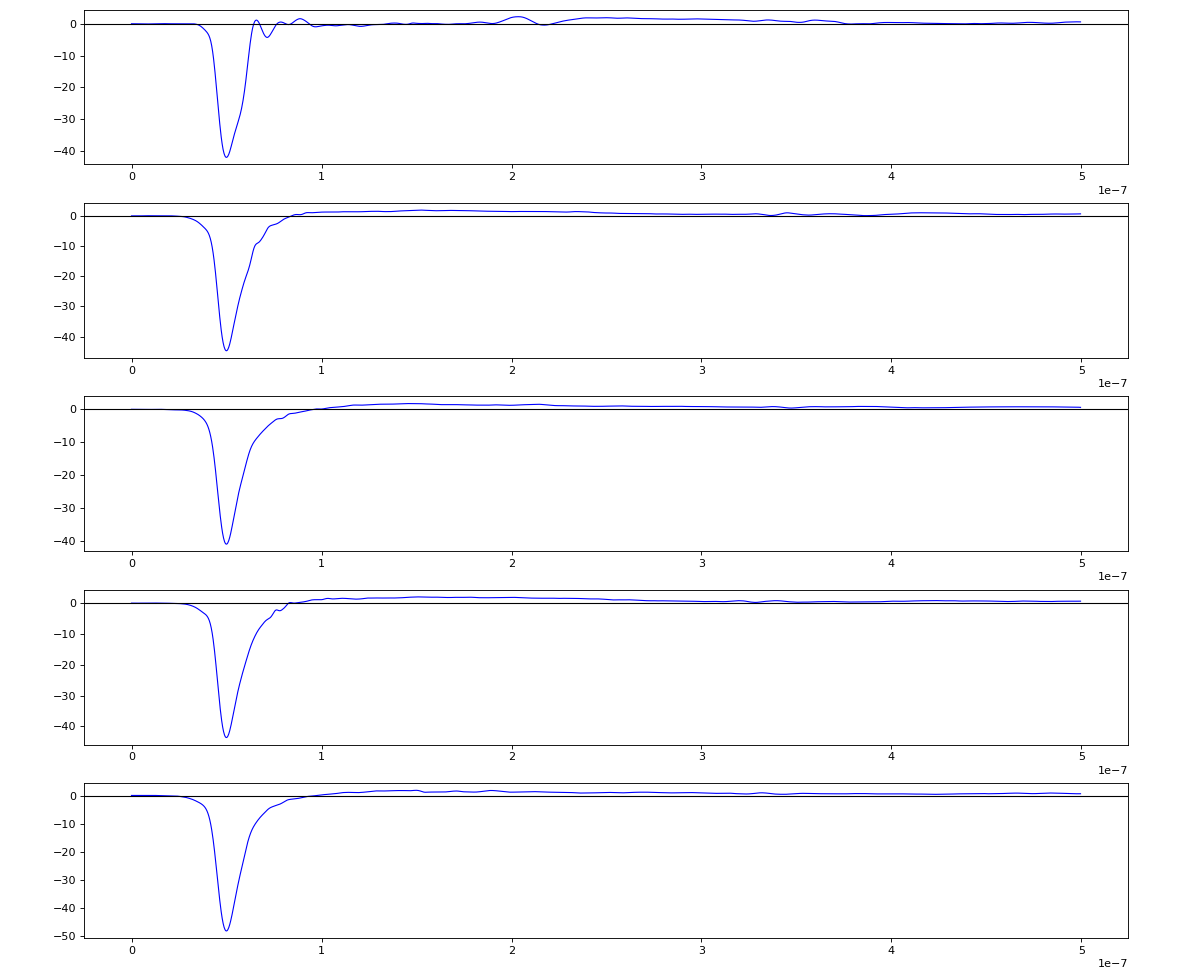
Wavelets estimation
Waveform per channel averaged over 1k raw events with selection of a single peak for amox27716 runs 91,92, and 93
Calibration
Updated on 2020-10-08
Current hex-/quad-anode calibration procedure is not automated, and needs in expert manual operations...
Example scripts
- example scripts reside in the directory
lcls2/psana/psana/hexanode/examples/ - calibration of quad-anode requires two passes over ~105 events data set. There are two different approaches for calibration;
- slow calibration - run script like
ex-24-quad-proc-sort-graph.pytwo times directly on xtc2 data file for waveform processing and calibration. - faster calibration (x2 faster, especially helpful at development) - run script like
ex-22-data-acqiris-peaks-save-h5.pyorex-22-data-acqiris-peaks-save-h5-xiangli.pyonce on xtc2 data file for waveform processing and save intermediate results for number of hits per channel and times per channel in compact hdf5 file.- run script like
ex-23-quad-proc-sort-graph-from-h5.pytwo times on hdf5 data file for calibration.
- run script like
- slow calibration - run script like
- Approach 2 is used for further example because compact hdf5 file with 60k events is available in
/reg/g/psdm/detector/data_test/hdf5/amox27716-r0100-e060000-single-node.h5, but real xtc2 data files are not available yet.
What needs to be done in two calibration passes
Calibration pass 1
- edit file copied from
hexanode/examples/configuration_quad.txt- set the 1-st parameter "command" to 2.
make sure that the beginning 10 lines of this script are consistent with what you are doing, e.g.
2 // -1 = detector does not exist, 0 = just read (no sort/calib), 1 = sort, 2 = calibrate fu,fv,fw, w_offset, 3 = generate correction tables and write them to disk 0 // hexanode used (yes = 1, no = 0) (Parameter 1101) 0 // 0 = common start, 1 = common stop (for TDC8HP and fADC always use 0) 1 // TDC channel for u1 (counting starts at 1) (Parameter 1129) 2 // TDC channel for u2 (counting starts at 1) (Parameter 1130) 3 // TDC channel for v1 (counting starts at 1) (Parameter 1131) 4 // TDC channel for v2 (counting starts at 1) (Parameter 1132) 0 // TDC channel for w1 (counting starts at 1) (Parameter 1133) HEX ONLY 0 // TDC channel for w2 (counting starts at 1) (Parameter 1134) HEX ONLY 5 // TDC channel for mcp(counting starts at 1) (0 if not used) (Parameter 1135)
- in the script like
ex-23-quad-proc-sort-graph-from-h5.pyset path to the fileconfiguration_quad.txte.g.' calibcfg' : '/reg/neh/home4/dubrovin/LCLS/con-lcls2/lcls2/psana/psana/hexanode/examples/configuration_quad.txt', - After event loop is completed, this script generates a lot of histograms (BTW output histograms are controlled by the kwargs dict, see ex-23*).
- Look at resulting histograms and edit
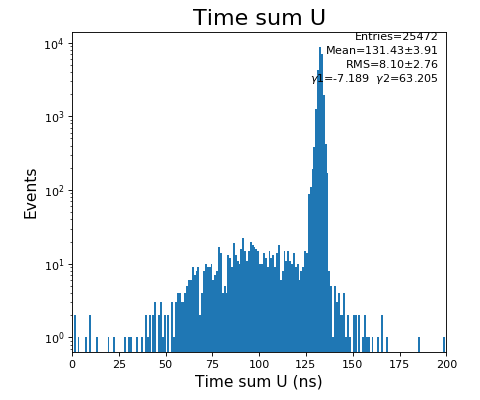
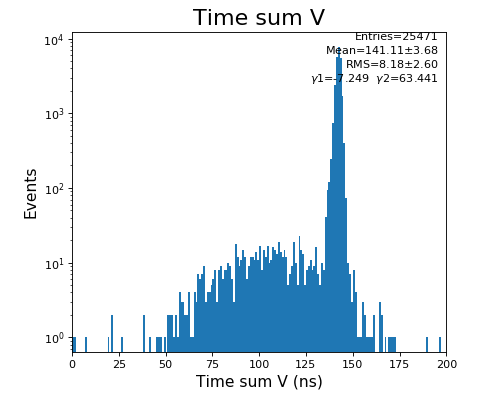
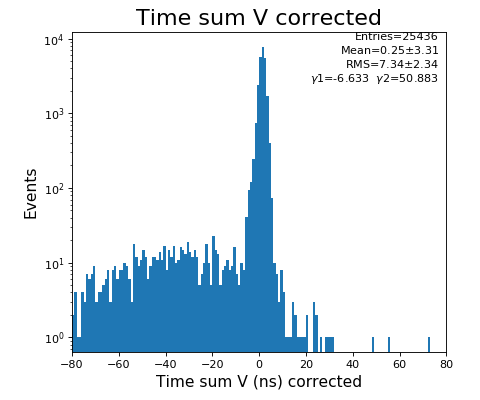
configuration_quad.txt- use Time sum U and V plots to set peak time value in Parameter 1108,1109
- then Halfwidth of the peak in this distribution to set Parameter 1115, 1116
- then Halfwidth of the u(ns), v(ns) distribution to set Parameter 1118, 1119
- manually set scale-factor for layer U and V (mm/ns) Parameter 1102, 1103
Calibration pass 2
- edit file
configuration_quad.txtand set the 1-st parameter "command" to 3. - edit script like
ex-23-quad-proc-sort-graph-from-h5.pyand set pass where time correction table will be saved, e.g.'calibtab' : '/reg/neh/home4/dubrovin/LCLS/con-lcls2/lcls2/psana/psana/hexanode/examples/calibration_table_data_new.txt' - run again script
ex-23-quad-proc-sort-graph-from-h5.py - check results in
calibration_table_data_new.txt
Post calibration arrangements
- edit file
configuration_quad.txt- set the 1-st parameter "command" to 1.
- set Parameter 1124 in configuration_quad.txt
Both, configuration and calibration files need to be saved in calibration DB for automated data processing. This operation needs in actual experiment and detector names, run, timestamp, etc. Preliminary deployment commands:
cdb add -e amox27716 -d tmo_quadanode -c calibcfg -r 100 -f configuration_quad.txt -i txt -u dubrovin cdb add -e amox27716 -d tmo_quadanode -c calibtab -r 100 -f calibration_table_data.txt -i txt -u dubrovin
Content of the DB can be explored with GUI started by command
calibman.
Automated data processing
- See examples
ex-24*orex-25*.
Plots
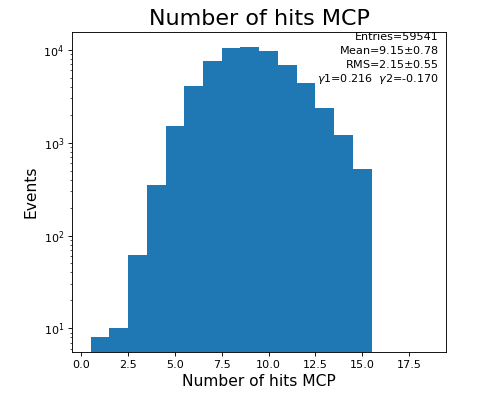
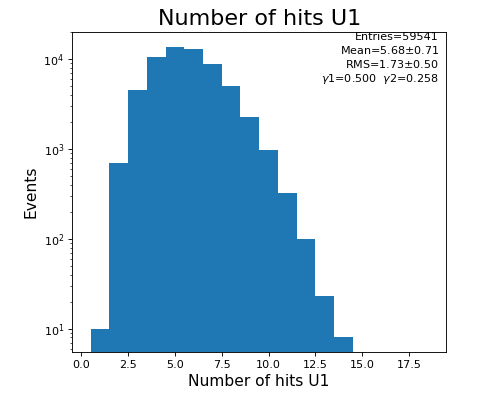
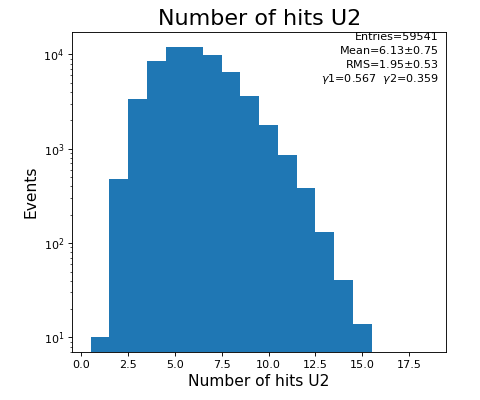
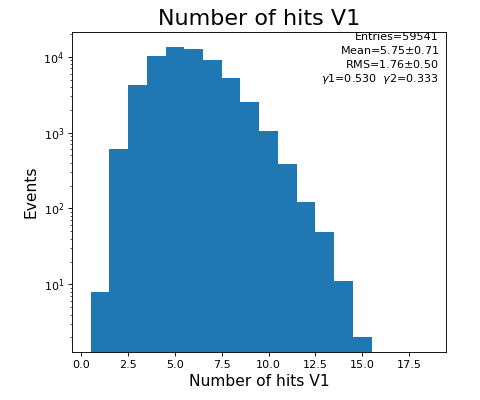
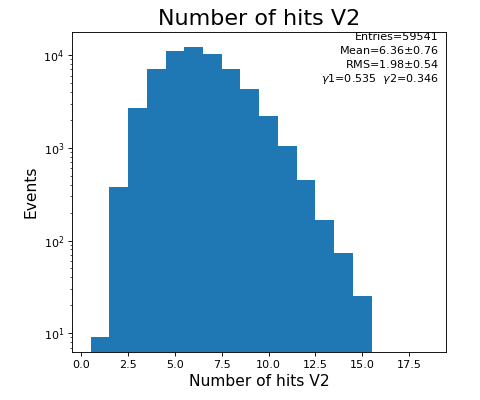
Number of hits per channel
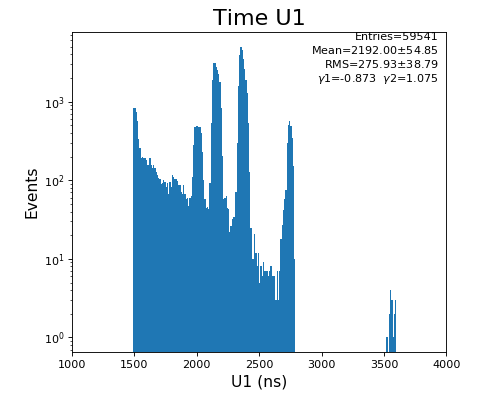
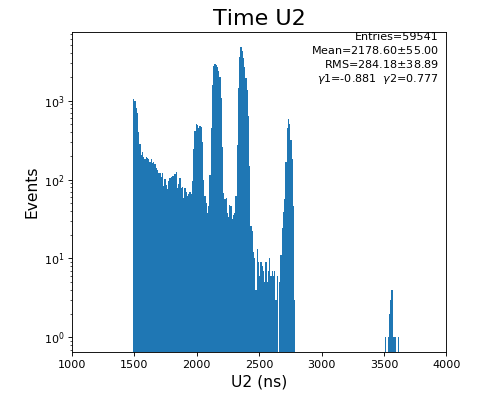
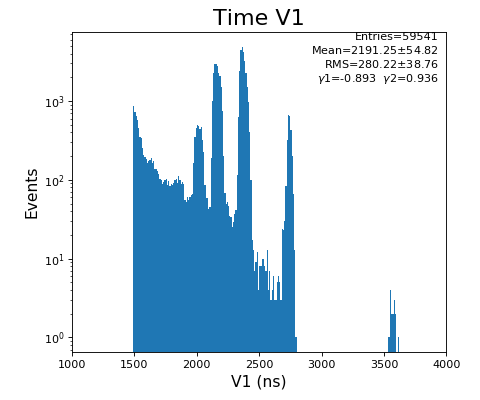
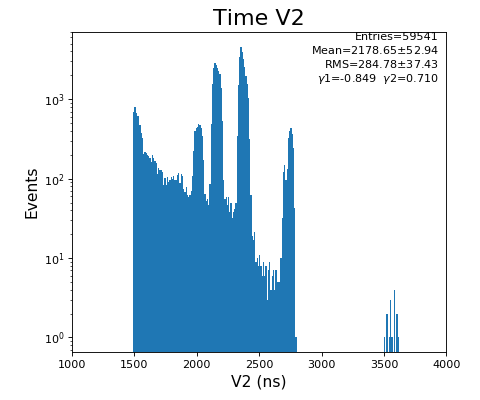
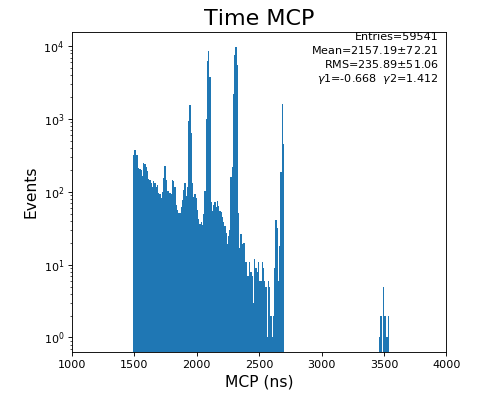
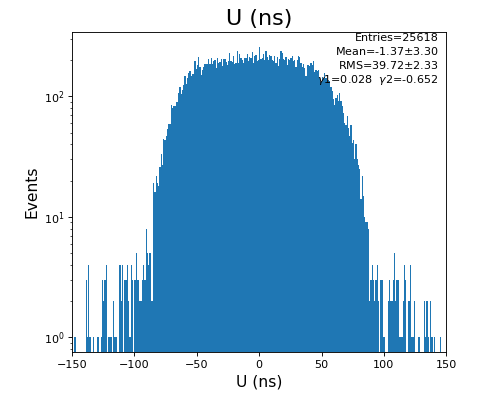
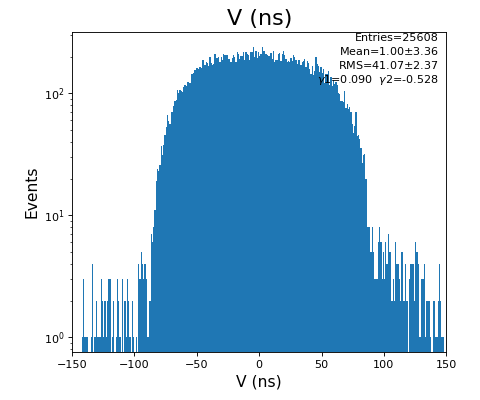
Spectra of time per channel, Spectra of U, V (ns)
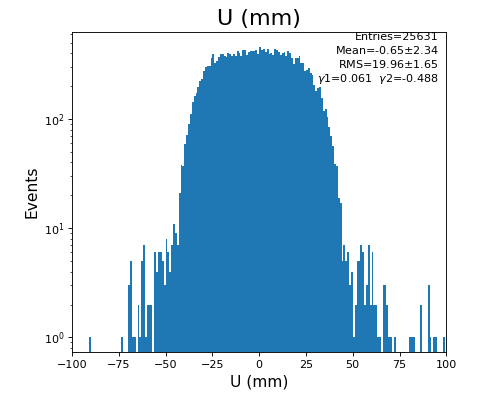
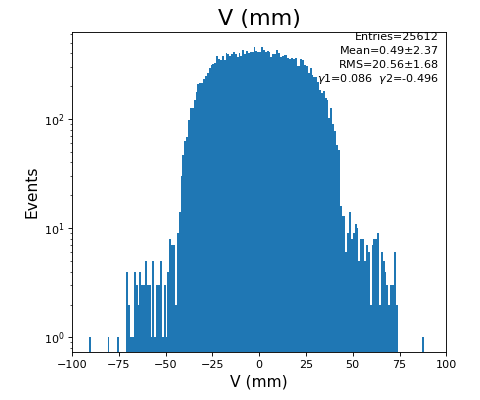
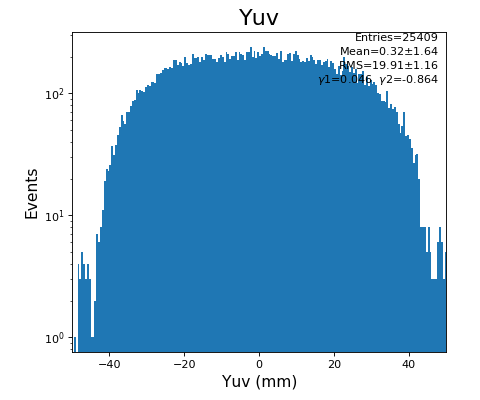
Spectra of U, V (mm)
Time sum (ns) for U, V
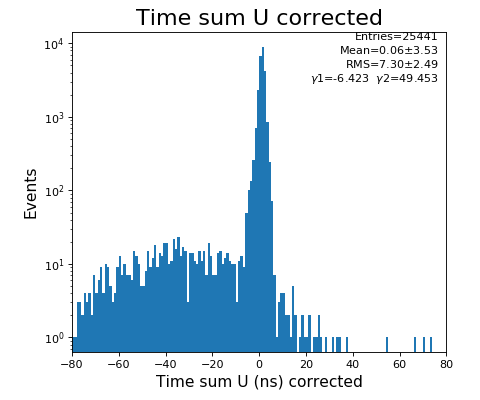
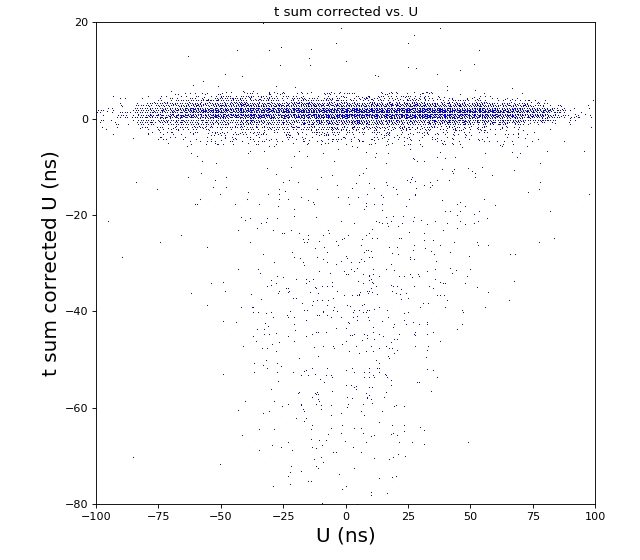
Time sum (ns) corrected for U, V
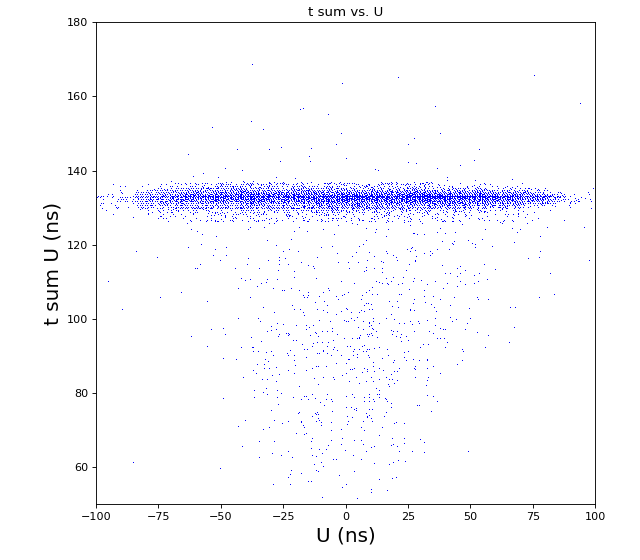
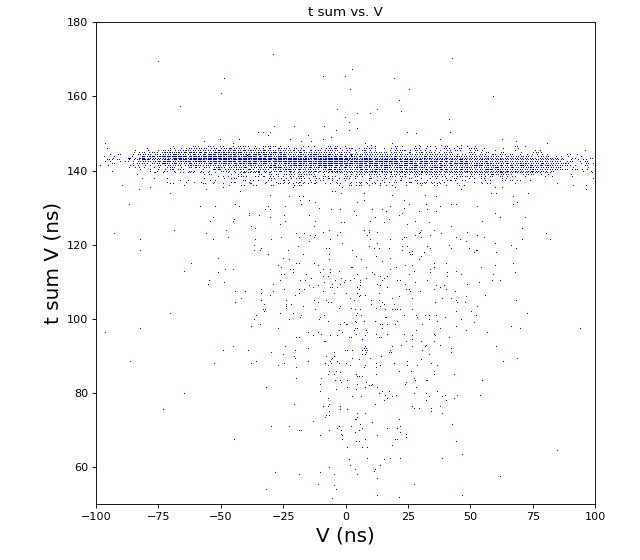
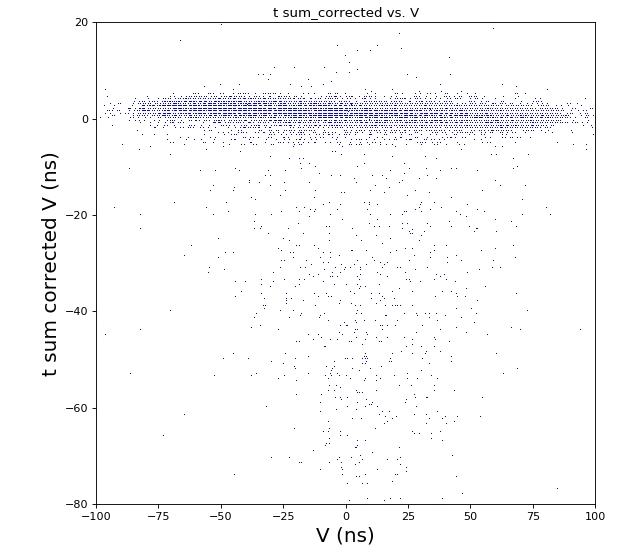
Time sum vs. variable U, V
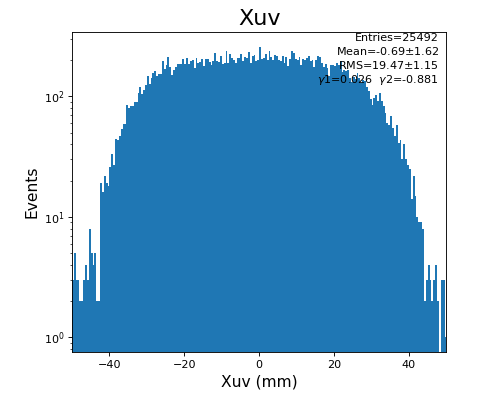
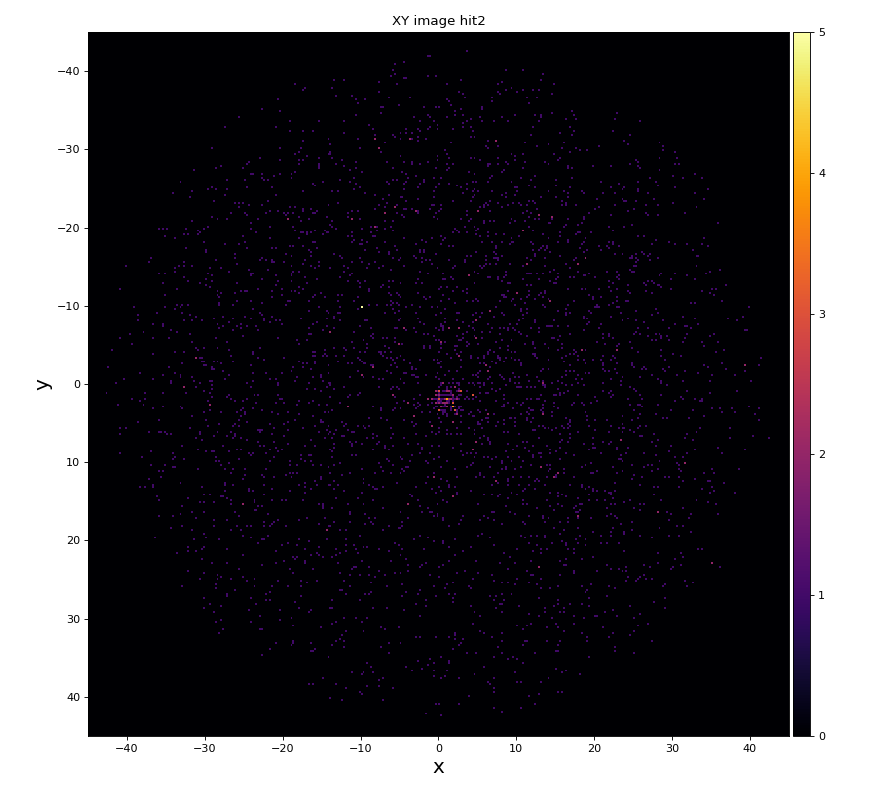
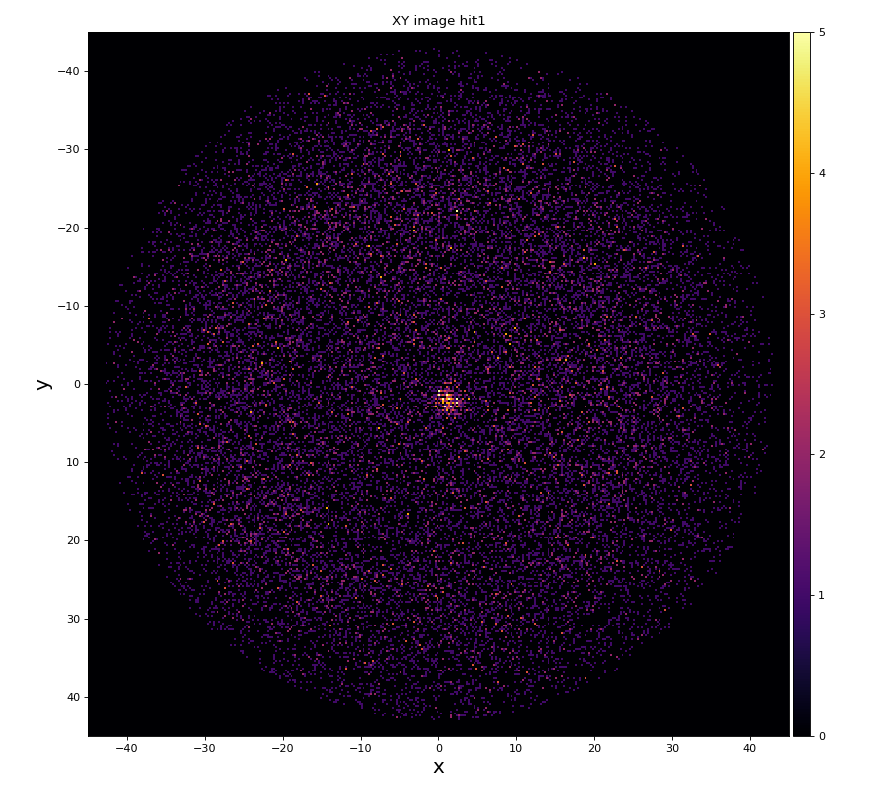
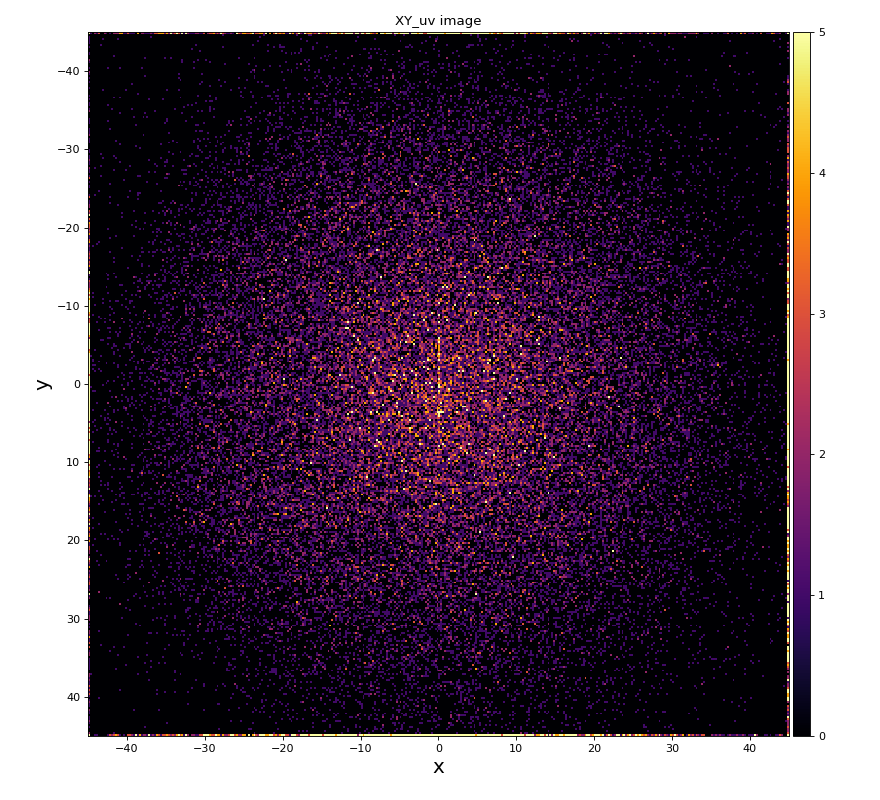
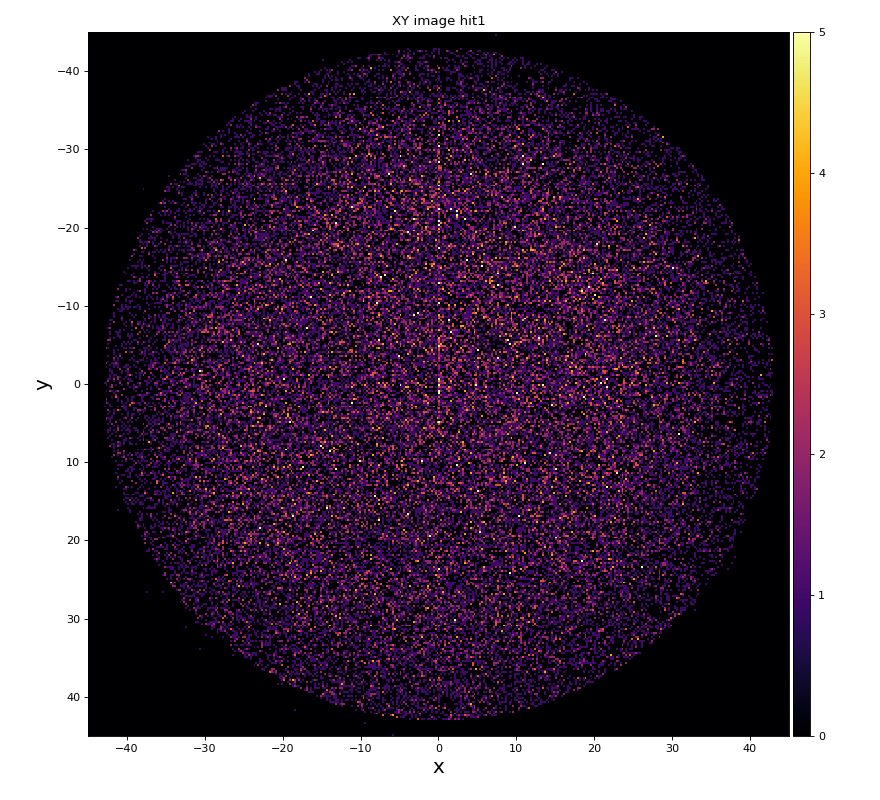
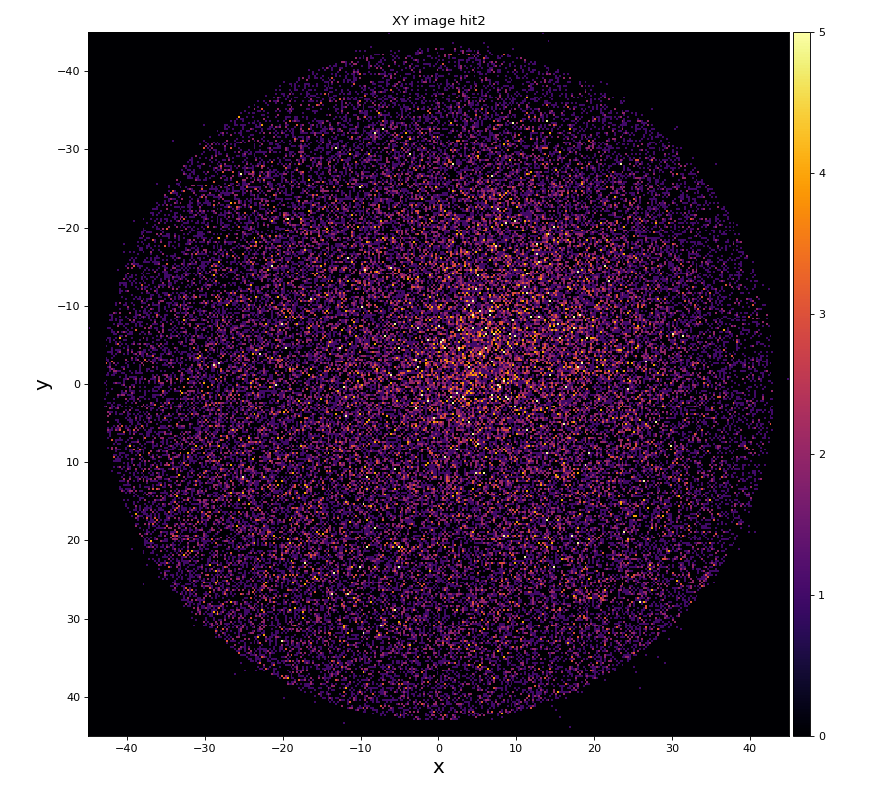
xy image for hit1 and 2
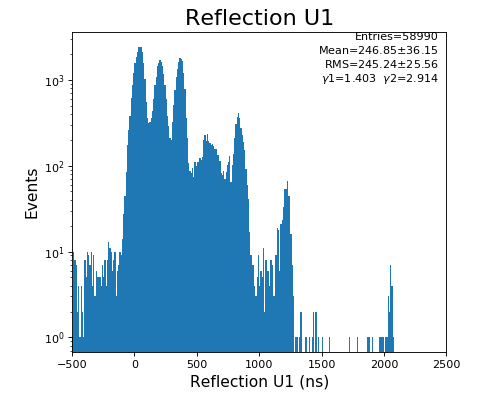
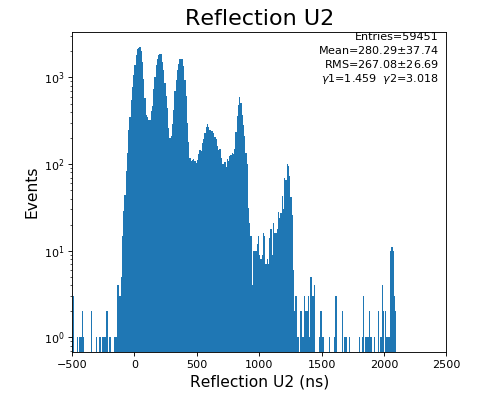
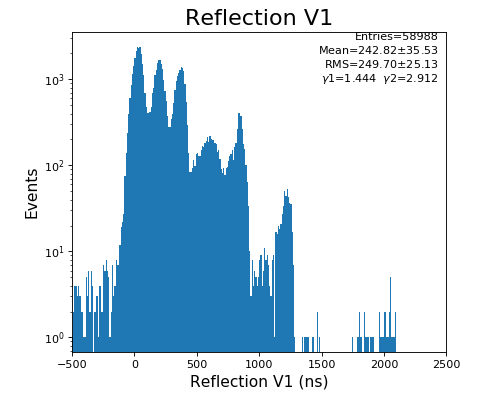
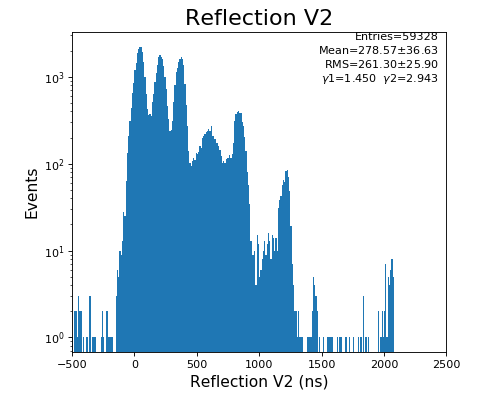
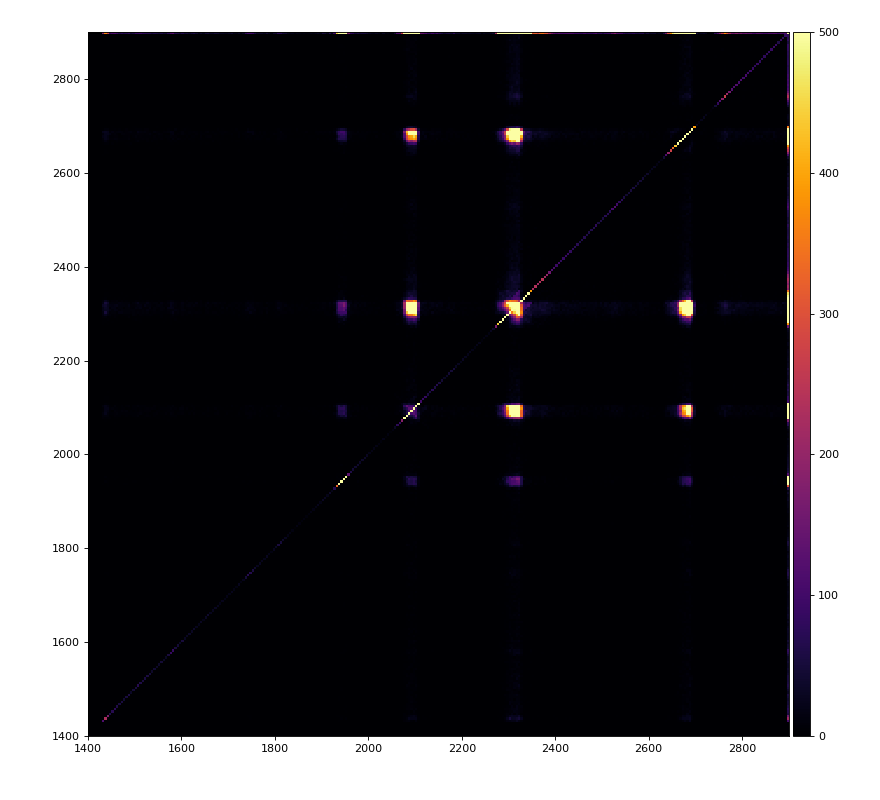
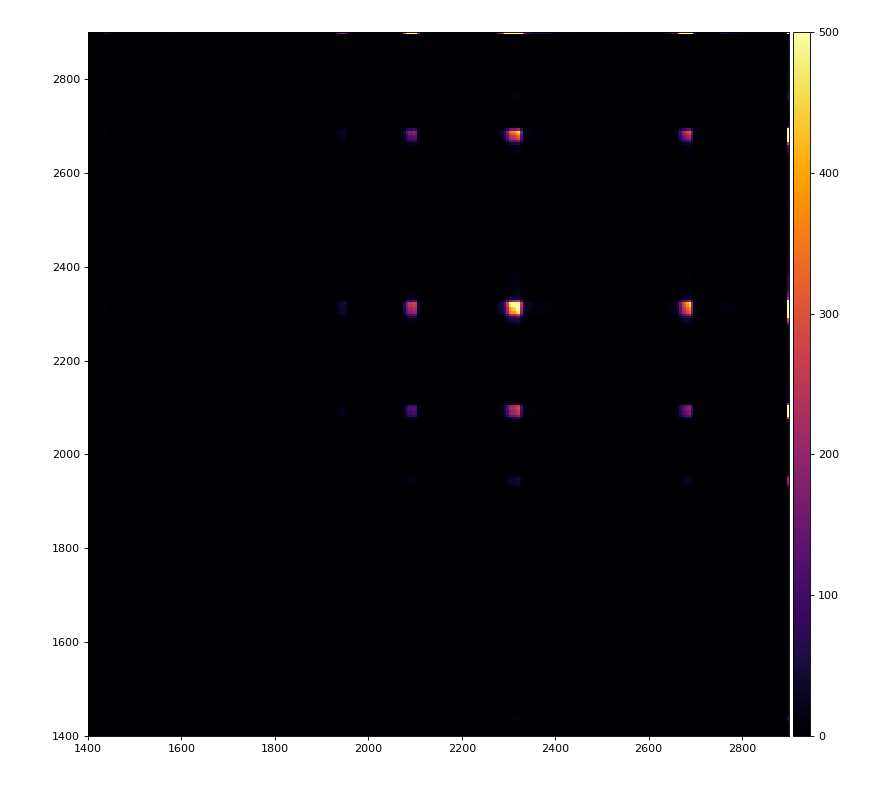
Reflection for all channels
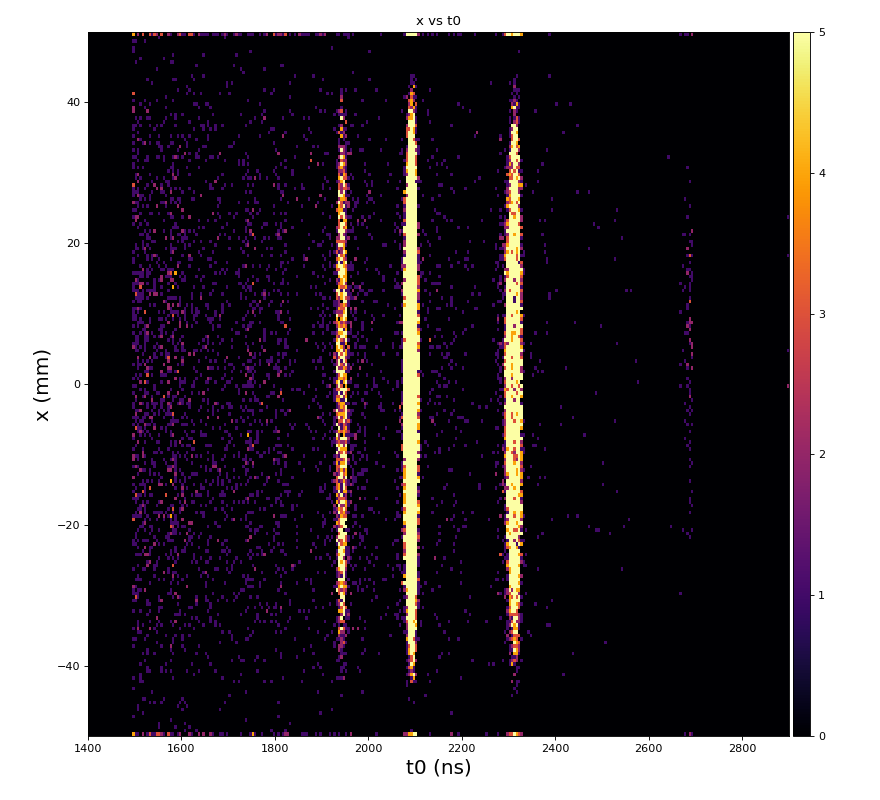
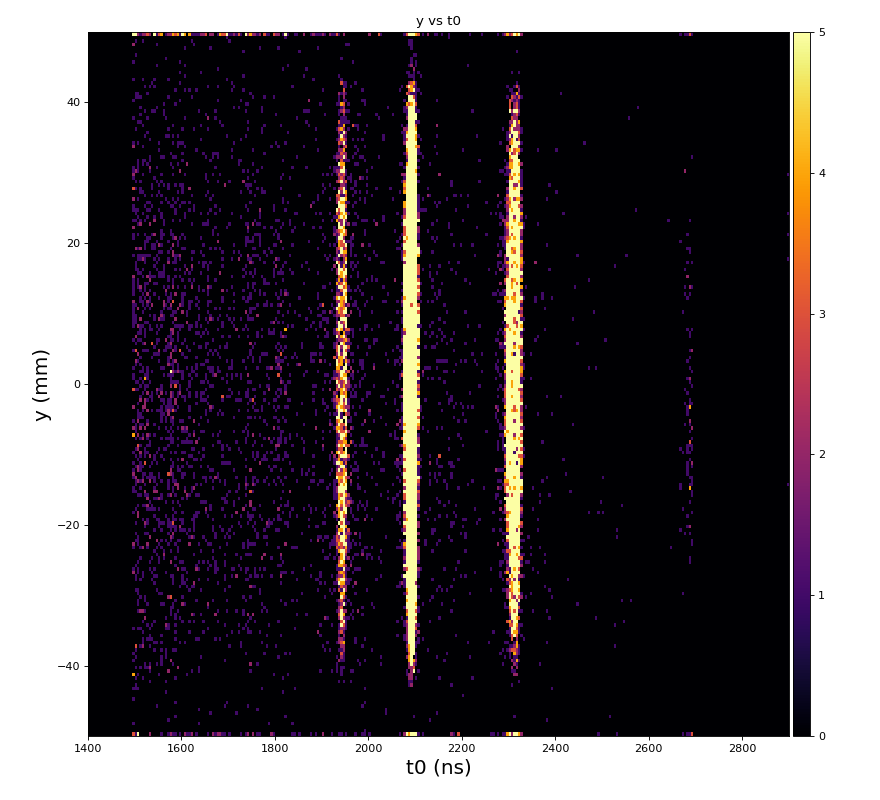
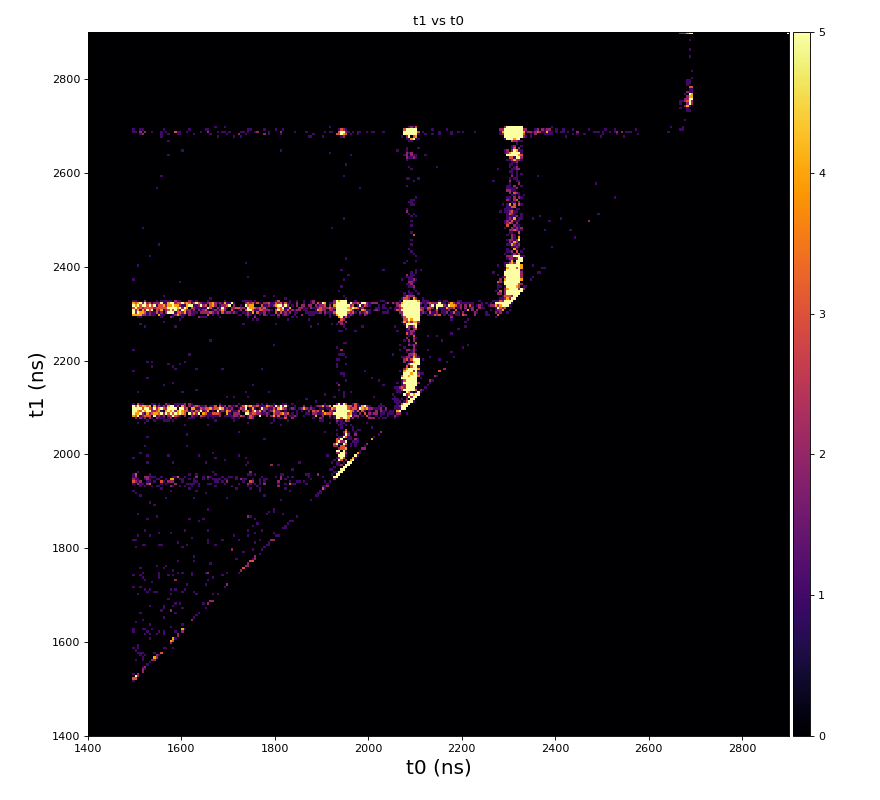
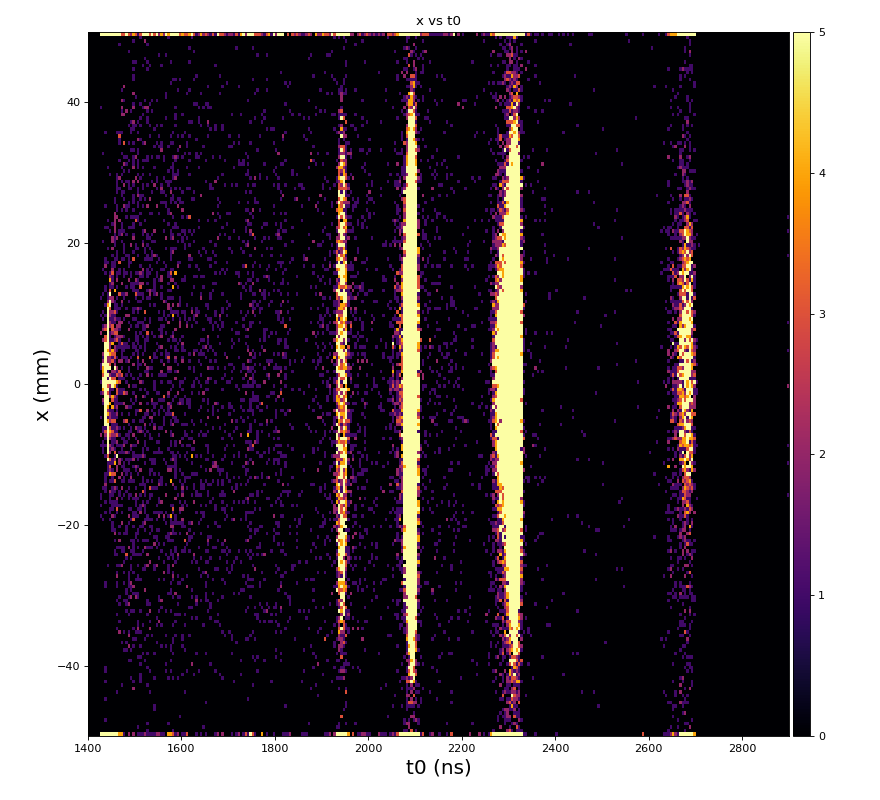
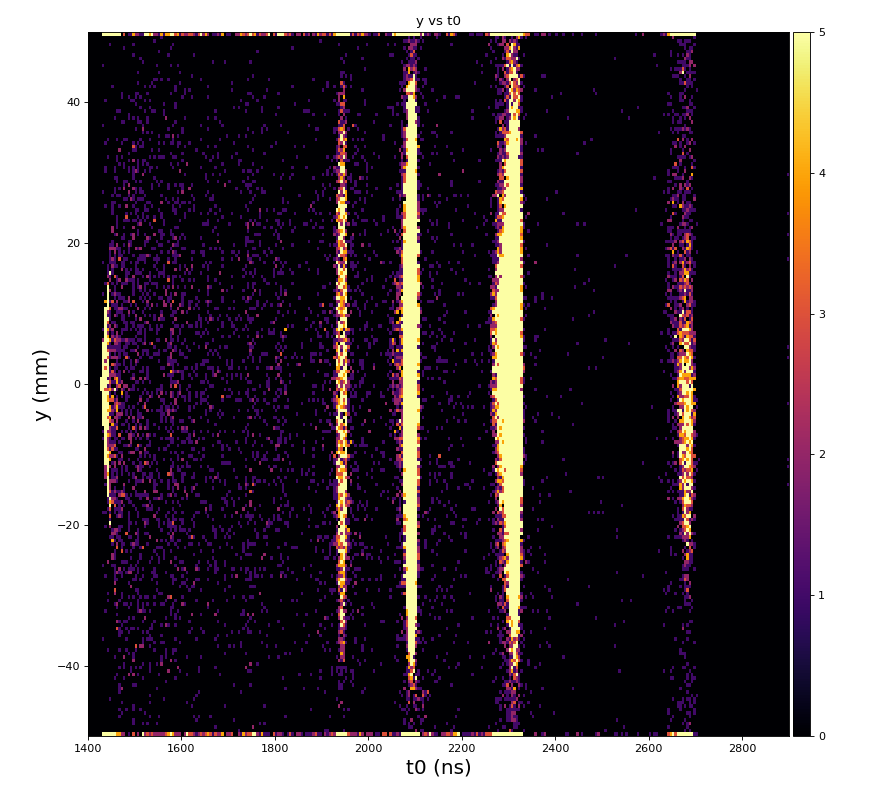
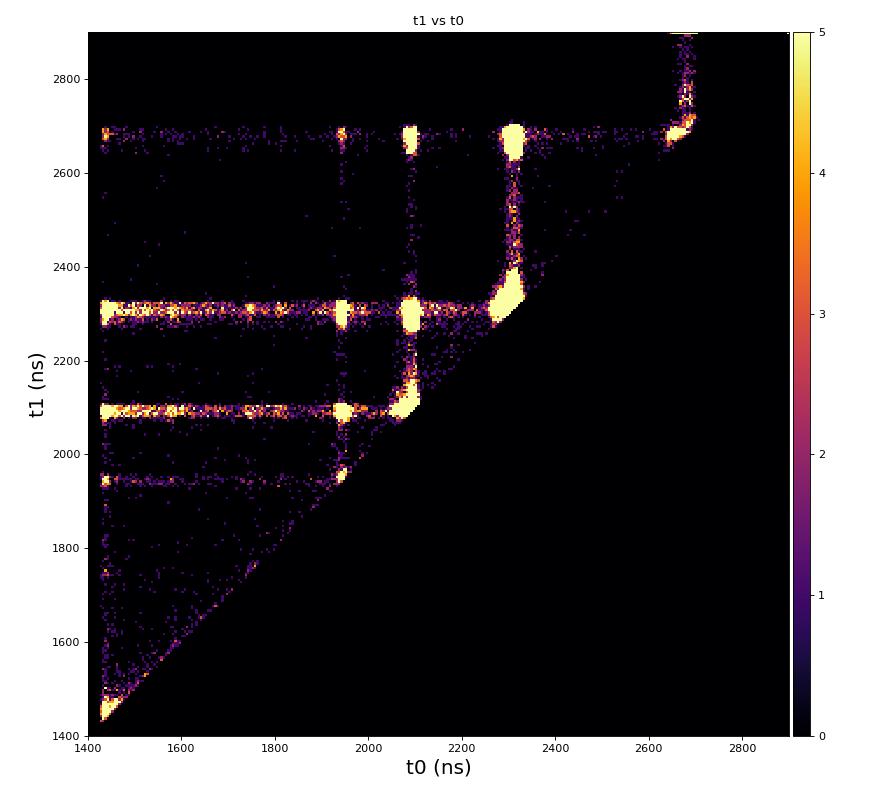
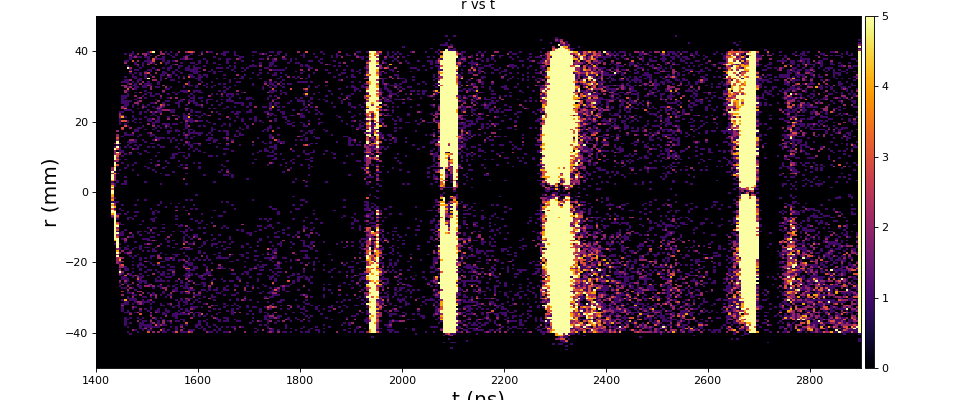
Physics plots t1,x,y vs t0
Calibrated plots
hexanode/examples/ex-quad-09-sort-graph-data.py with command 1 (after calibration command 2,3) set in configuration_quad.txt.
Soft on LCLS2
Examples
https://github.com/slac-lcls/lcls2/blob/master/psana/psana/hexanode/examples/
- ex-20-data-acqiris-access.py - access to detector waveforms
- ex-21-data-acqiris-graph.py - plot waveforms and found peaks
- ex-22 - 24 - intended for calibration and representative graphics
- ex-25-quad-proc-data.py - reads waveforms from xtc2, find peaks, and reconstruct hits using Roentdec library
Physics plots
From ex-23-quad-proc-sort-graph-from-h5.py or ex-24 if xtc2 file is available:
XTC2 trick
Only useful channels will be selected in xtc2. This hack is temporary available in
psana/psana/detector/test_detectors.py
References
- Hexanode detector test on data
- Hexanode users' examples
- TMO data flow spread sheet
- Covariance mapping
- 2019-11-01-Peter-Walter-article.pdf