Page History
Policies
LCLS users are responsible for complying with the data management and curation policies of their home institutions and funding agents and authorities. To enhance the scientific productivity of the LCLS user community, LCLS supplies on-site disk, tape and compute resources for prompt analysis of LCLS data, and software to access those resources consistent with the published data retention policy. Compute resources are preferentially allocated to recent and running experiments.
Data Management
LCLS provides space for all your experiment's data at no cost for you. This includes the raw data from the detectors as well as the data derived from your analysis. Your raw data are available as XTC files or, on demand, as HDF5 files. The tools for managing files are described here
Getting an Account
You will need a valid SLAC UNIX account in order to use the LCLS computing system. The instructions for getting a SLAC UNIX account are here:
http://www-ssrl.slac.stanford.edu/lcls/users/logistics.html#compaccts
Your UNIX account must be enabled in the LCLS system in order to have access to data and elog. This happens automatically if your account is created with XU as its primary group. If your primary UNIX group is not XU, make a request of enabling your account in the LCLS system by sending an email to:
If you forgot your password or if your account has been disabled send an email to:
account-services@slac.stanford.edu
Getting Access to the System
You can get into the LCLS photon computing system by ssh'ing to:
| No Format |
|---|
psexport.slac.stanford.edu
|
From these nodes you can move data files in and out of the system and you can connect to the bastion hosts:
| No Format |
|---|
pslogin
|
Note that, from within SLAC, you can directly connect to the bastion hosts without going through psexport.
The SLAC wireless visitor network is not considered part of SLAC so you'll need to go through psexport when using your laptop on-site.
From the bastion hosts you can then reach the analysis nodes (see below).
Each control room has a number of nodes for local login. These nodes have access to the Internet and are named psusr<id>.
The controls and DAQ nodes used for operating an instrument work in kiosk mode so you don't need a personal account to run an experiment from the control room. Remote access to these nodes is not allowed for normal users.
Running the Analysis
The analysis framework is documented in the Data Analysis page for the LCLS-I/HXR systems and psana for the LCLS-II (SXR&UED) systems. This section describes the nodes which are resources available for running the analysis.
Interactive Pools
In order to get access to the interactive nodes, connect to the addresses psananeh or psanafeh. A load-balancing mechanism will connect you to the least loaded of the nodes in the pool:
| No Format |
|---|
ssh psananeh
ssh psanafeh
|
Each pool is currently made of six servers with the following general specifications:
- 8-cores, Opteron 2384, 8GB, diskless, 10Gb/s
Each node in the interactive pools has one single user Matlab license. You can find which nodes in the pool have a Matlab license available by running the following command on any of the psana nodes:
| No Format |
|---|
/reg/common/package/scripts/matlic
|
Batch Farm
There are batch farms located in the NEH and FEH. Depending on your data access you may need to submit jobs to a specific farm. This can be accomplished by submitting to the appropriate LSF batch queue. Refer to the table below. Multi-core OpenMPI jobs should be run in either the psnehmpiq or psfehmpiq batch queue, see the following section on "Submitting OpenMPI Batch Jobs". Simulation jobs should be submitted to the low priority queues psfehidle and psfehidle.
Experimental Hall | Queue | Nodes | Data | Comments |
|---|---|---|---|---|
NEH | psnehq | psana11xx,psana12xx | ana01, ana02 | Jobs <= 6 cores |
| psnehmpiq | psana11xx,psana12xx | ana01, ana02 | OpenMPI jobs > 6 cores, preemptable |
| psnehidle | psana11xx,psana12xx |
| Simulations, preemptable, low priority |
FEH | psfehq | psana13xx,psana14xx | ana11, ana12 | Jobs <= 6 cores |
| psfehmpiq | psana13xx,psana14xx | ana11, ana12 | OpenMPI jobs > 6 cores, preemptable |
| psfehidle | psana13xx,psana14xx |
| Simulations, preemptable, low priority |
You can find more LCLS specific information about LSF in this PDF file. For a more detailed description and more LSF commands, please see:
http://www.slac.stanford.edu/comp/unix/unix-hpc.html
The batch farm is made of eighty servers with the following general specifications:
- 12 cores (24 with Hyperthreading), Xeon X5675, 24GB memory, 500GB disk, QDR IB
Submitting Batch Jobs
Login first to pslogin (from SLAC) or to psexport (from anywhere). From there you can submit a job with the following command:
| No Format |
|---|
bsub -q psnehq -o <output file name> <job_script_command>
|
For example:
| No Format |
|---|
bsub -q psnehq -o ~/output/job.out my_program
|
This will submit a job (my_program) to the queue psnehq and write its output to a file named ~/output/job.out.
You may check on the status of your jobs using the bjobs command.
Submitting OpenMPI Batch Jobs
The RedHat supplied OpenMPI packages are installed on pslogin, psexport and all of the psana batch servers.
The system default has been set to the current version as supplied by RedHat.
| No Format |
|---|
$ mpi-selector --query
default:openmpi-1.4-gcc-x86_64
level:system
|
Your environment should be set up to use this version (unless you have used RedHat's mpi-selector script, or your login scripts, to override the default). You can check to see if your PATH is correct by issuing the command which mpirun. Currently, this should return /usr/lib64/openmpi/1.4-gcc/bin/mpirun. Future updates to the MPI version may change the exact details of this path.
In addition, your LD_LIBRARY_PATH;should include /usr/lib64/openmpi/1.4-gcc/lib (or something similar).
For notes on compiling examples; please see:
http://www.slac.stanford.edu/comp/unix/farm/mpi.html
The following are examples of how to submit OpenMPI jobs to the PCDS psnehmpiq batch queue:
| No Format |
|---|
bsub -q psnehmpiq -a mympi -n 32 -o ~/output/%J.out ~/bin/hello
|
Will submit an OpenMPI job (-a mympi) requesting 32 processors (-n 32) to the psnehmpiq batch queue (-q psnehmpiq).
| No Format |
|---|
bsub -q psfehmpiq -a mympi -n 16 -R "span[ptile=1]" -o ~/output/%J.out ~/bin/hello
|
| Wiki Markup |
|---|
Will submit an OpenMPI job (-a mympi) requesting 16 processors (-n 16) spanned as one processor per host (-R "span\[ptile=1\]") to the psfehmpiq batch queue (-q psfehmpiq). |
| No Format |
|---|
bsub -q psfehmpiq -a mympi -n 12 -R "span[hosts=1]" -o ~/output/%J.out ~/bin/hello
|
| Wiki Markup |
|---|
Will submit an OpenMPI job (-a mympi) requesting 12 processors (-n 12) spanned all on one host (-R "span\[hosts=1\]") to the psfehmpiq batch queue (-q psfehmpiq). |
Data Storage
LCLS provides space for all your experiment's data at no cost for you. This includes the measurements as well as the data derived from your analysis. Your raw data are available as XTC files or, on demand, as HDF5 files. The LCLS data policy is described here. The path name of the experiment data is /reg/d/psdm.
Data Export
There is a web interface to the experimental data accessible via
https://pswww.slac.stanford.edu/apps/explorer
The web interface also allows you to generate file lists that can be fed to the tool you use to export the data from SLAC to your home institution. You can use psexport for copying your data.
The recommended tools for exporting the data offsite are bbcp and Globus Online. The former, bbcp, is slightly simpler to setup. On the other hand Globus Online is more efficient when transferring large amount of data because it babysits the overall process by, for example, automatically restarting a failed or stalled transfer. The performance of the two tools is very similar.
Printing
The following printers are available in the NEH building from all the UNIX nodes:
...
Info
...
Location
...
Device URI
...
Dell 3130
...
AMO Control Room
...
lpd://dellcolor-neh-amo1/lp
...
Dell 3130
...
AMO Control Room
...
lpd://dellcolor-neh-amo2/lp
...
Dell 3130
...
SXR Control Room
...
lpd://dellcolor-neh-sxr1/lp
...
Dell 3130
...
SXR Control Room
...
lpd://dellcolor-neh-sxr2/lp
...
Dell 3130
...
XPP Control Room
...
lpd://dellcolor-neh-xpp1/lp
...
Dell 3130
...
XPP Control Room
...
lpd://dellcolor-neh-xpp2/lp
...
HP Color LaserJet CP3525
...
Bldg 950 corridor ground floor
...
ipp://hpcolor-neh-corridor/ipp/
...
Xerox WorkCentre 5675
...
Bldg 950 Rm 218, Jason Alpers
...
ipp://hpcolor-neh-laser/ipp/
...
HP Color LaserJet 4700
...
Bldg 950 Rm 204, Ray Rodriguez
...
ipp://hpcolor-neh-ray/ipp/
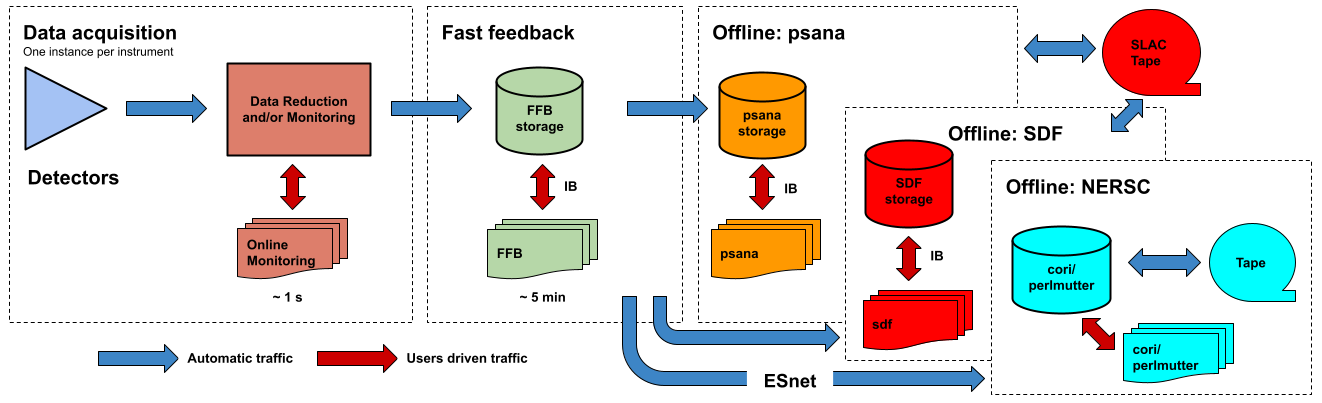
The following figure shows a logic diagram of the LCLS data flow and indicates the different stages where data analysis can be performed in LCLS:
- Data acquisition - The online monitoring nodes get detector data over the network and place it in shared memory on each node. There is a set of monitoring nodes allocated for each instrument. The detector data are received over the network by snooping the multicast traffic between the DAQ readout nodes and the data cache layer. Analysis performed at this stage provides < 1 s feedback capabilities. The methods for doing (quasi) real time analysis are described in the Prompt Analysis page. Users should be aware of the different possibilities and choose the approach that works best for their experiment.
- Fast feedback - The processing nodes in the FFB system read the detector data from a dedicated, flash-based file system. It is possible to read the data as they are written to the FFB storage layer by the DAQ without waiting for the run to end. Analysis performed at this stage provides < 5 min feedback capabilities. The resources reserved for this stage are described in the Fast Feedback System page.
- Offline - The offline nodes read the detector data from disk. These systems include both interactive nodes and batch queues and are the main resource for doing data analysis. We currently support sending the data to three offline systems: psana, S3DF and NERSC. The psana system is the default offline system and your data will end up in psana unless you arrange a different destination with your experiment POC. The psana system is also relatively old and it will be retired when more storage becomes available in the S3DF system. Please consider running at NERSC if you expect to have intensive computing requirements (> 1 PFLOPS).
...
HP LaserJet 4350
...
Bldg 950 Rm 203
...