V3 News
- background shape is evaluated in run 162 WITHOUT common mode correction. See Masks, Background subtraction for detail.
- pfv3r1 - is upgraded PSAS-147
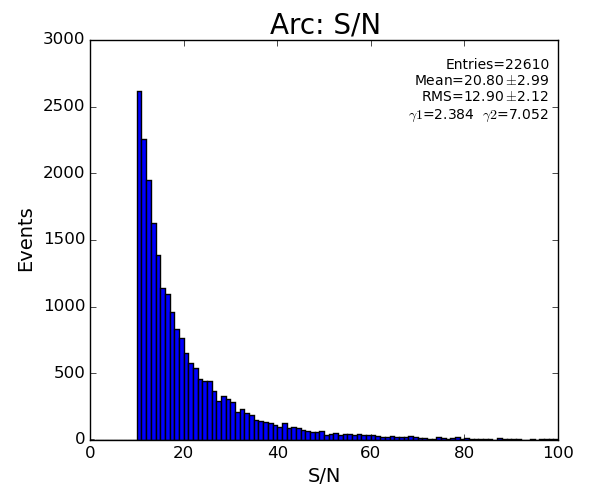
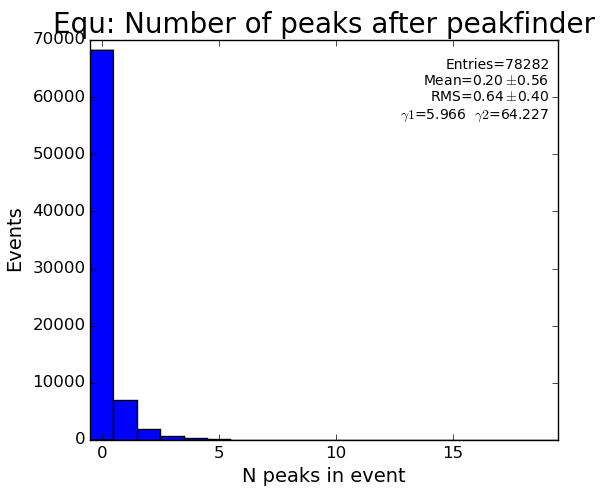
- S/N statistical significance is improved by increasing number of pixels. See Hit and Peak Finding Algorithms.
Data processing and peak finding is done in
cxif5315/proc-cxif5315-r0169-data-pfvn-2016-05-03.py- pfv3r1 was improved - now in the background avg and rms calculation discards all type of local maximums
Data processing
exp=cxif5315:run=169
nda_data = det.raw(evt)
if nda_data is not None :
nda = np.array(nda_data, dtype=np.float32, copy=True)
nda -= nda_peds
det.common_mode_apply(runnum, nda, cmpars=(5,50)) # applied for pfv3r1 latest processing only
nda = subtract_bkgd(nda, nda_bkgd, mask=nda_smask, winds=winds_bkgd, pbits=0)
nda *= nda_smask
Peak finder parameters
alg_arc = PyAlgos(windows=winds_arc, mask=mask_arc, pbits=2)
alg_equ.set_peak_selection_pars(npix_min=0, npix_max=1e6, amax_thr=0, atot_thr=0, son_min=10) # for v3r1
#alg_arc.set_peak_selection_pars(npix_min=0, npix_max=1e6, amax_thr=0, atot_thr=500, son_min=6) # for v2r1
#alg_arc.set_peak_selection_pars(npix_min=0, npix_max=1e6, amax_thr=0, atot_thr=0, son_min=6) # for v4r1
alg_equ = ... the same as above
# in the event loop
#peaks_arc = alg_arc.peak_finder_v2r1(nda, thr=30, r0=7, dr=2)
peaks_arc = alg_arc.peak_finder_v3r1(nda, rank=5, r0=7, dr=2)
#peaks_arc = alg_arc.peak_finder_v4r1(nda, thr_low=10, thr_high=150, rank=5, r0=7, dr=2)
peaks_equ = alg_equ... the same as above
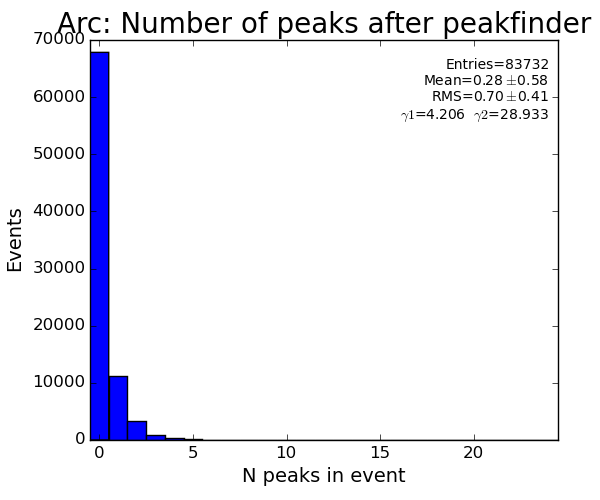
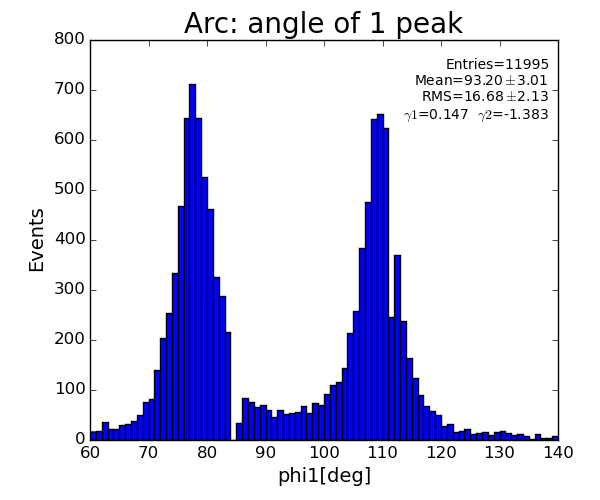
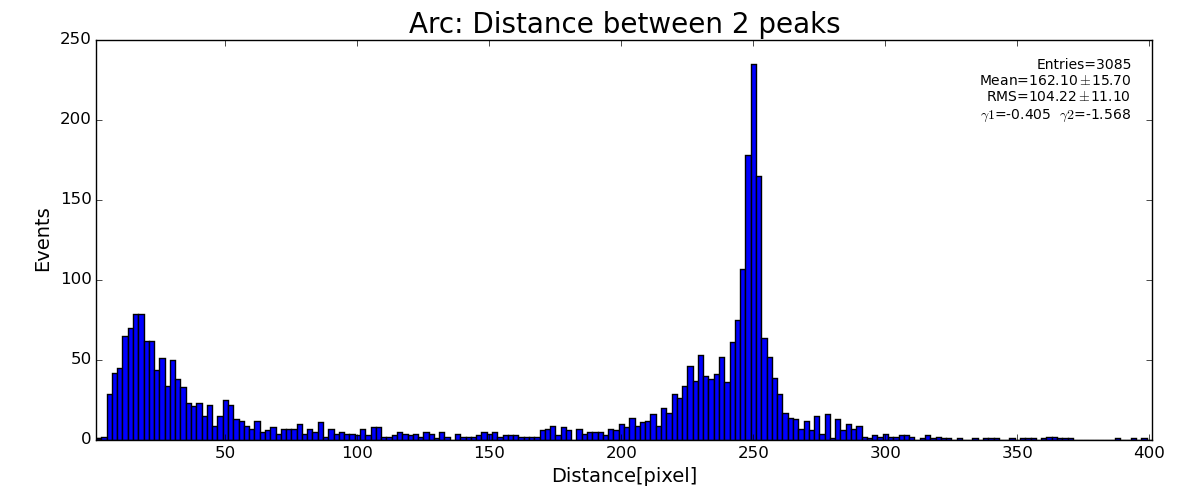
Results
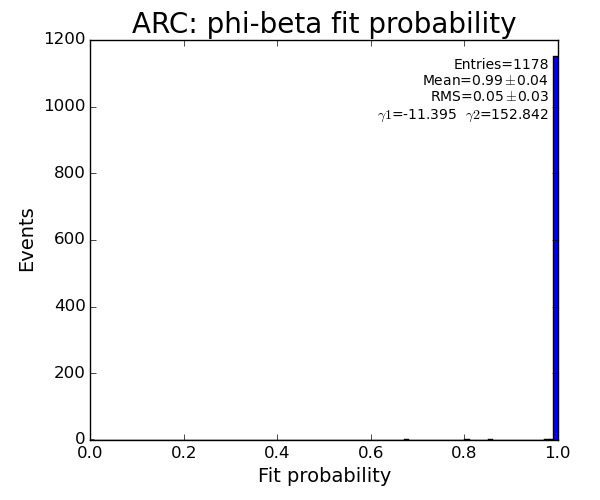
ARC
pre-selection for histograms
def procPeakDataArc(pk) :
""" Process peak for ARC region; accumulate peak statistics in histogram arrays.
"""
#===================
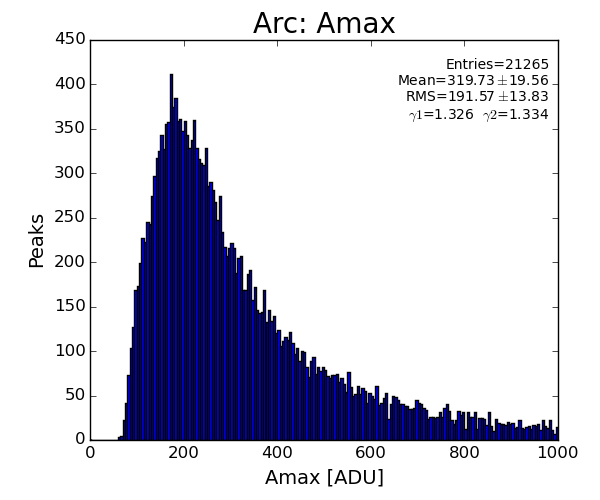
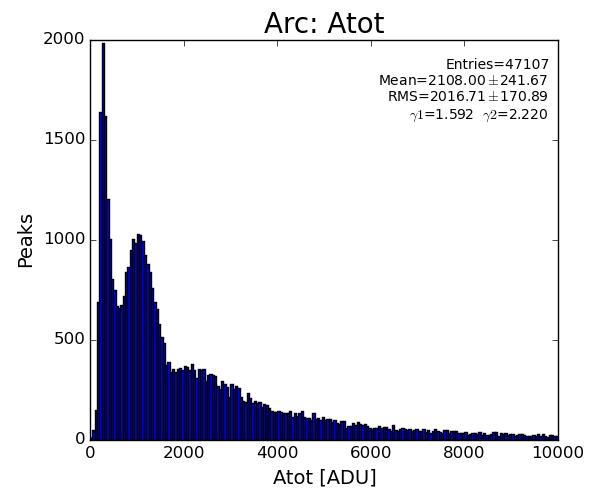
sp.lst_arc_atot.append(pk.atot)
if pk.atot<1500 : return
#===================
sp.lst_arc_amax.append(pk.amax)
sp.lst_arc_npix.append(pk.npix)
sp.lst_arc_r .append(pk.r)
sp.lst_arc_phi .append(pk.phi)
...
plots for pfv2/3/4r1
================
EQU
pre-selection for histograms
def procPeakDataEqu(pk) :
""" Process peak for EQU region; accumulate peak data
"""
# discard small radius peaks from all histograms
#===================
sp.lst_equ_atot.append(pk.atot)
if pk.atot<1500 : return
if pk.r<100 : return
#===================
sp.lst_equ_amax.append(pk.amax)
sp.lst_equ_npix.append(pk.npix)
sp.lst_equ_r .append(pk.r)
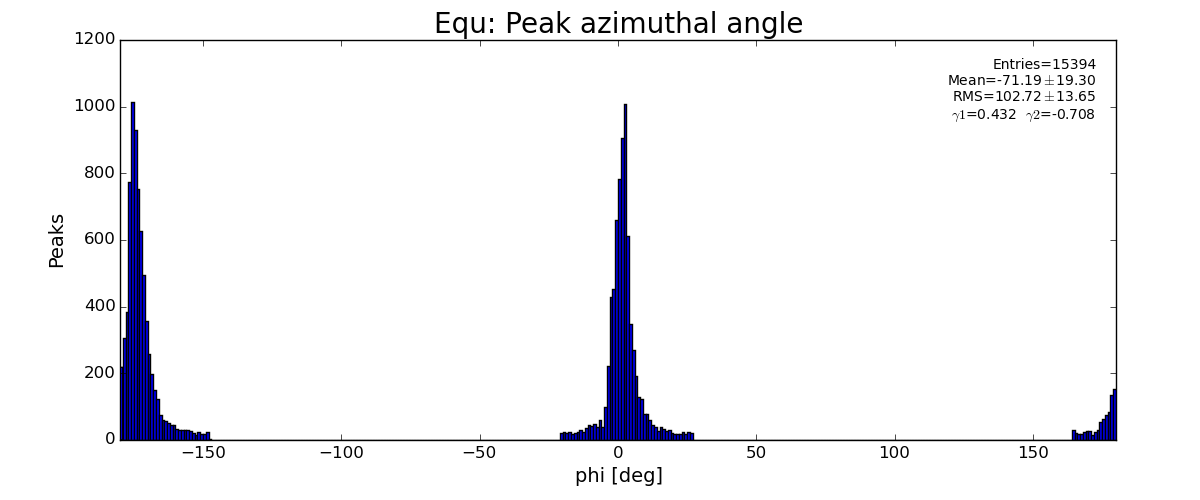
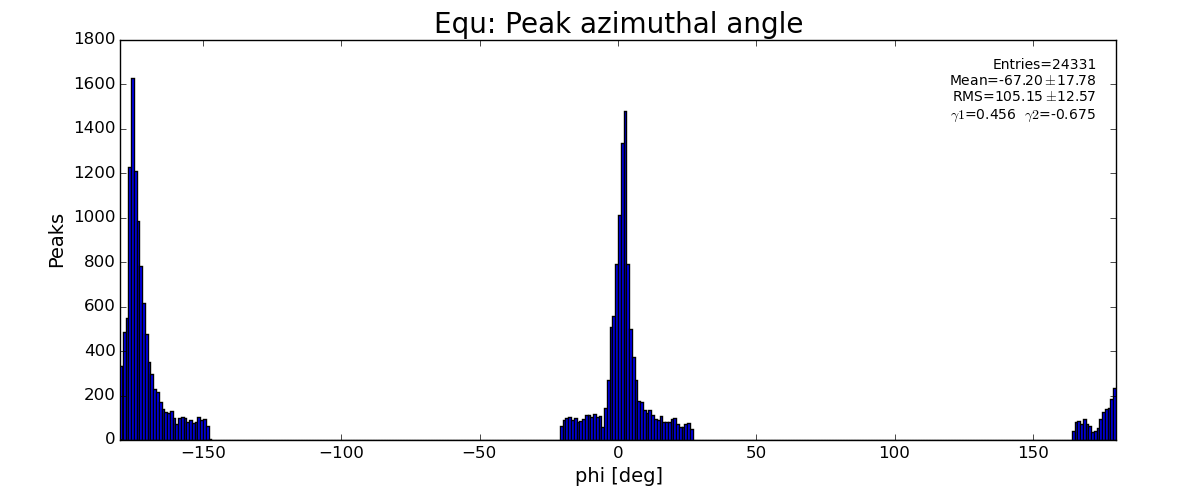
sp.lst_equ_phi .append(pk.phi)
...
plots for pfv2/3/4r1
================
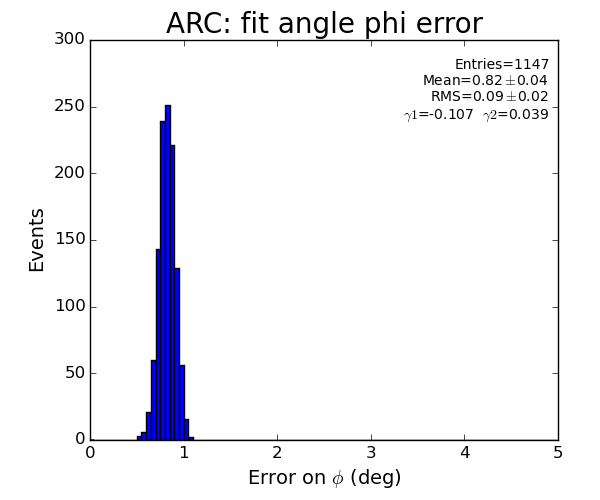
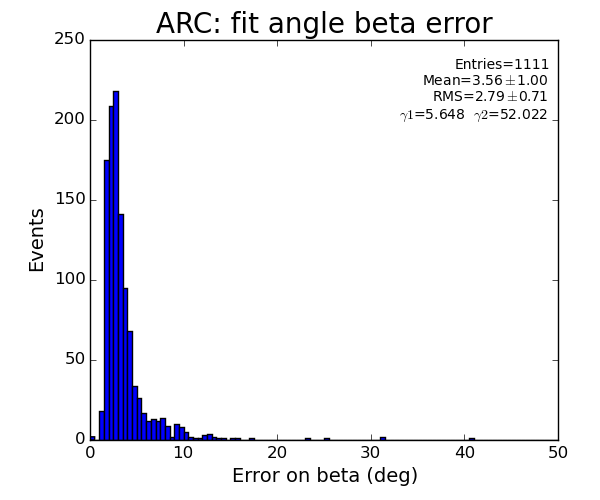
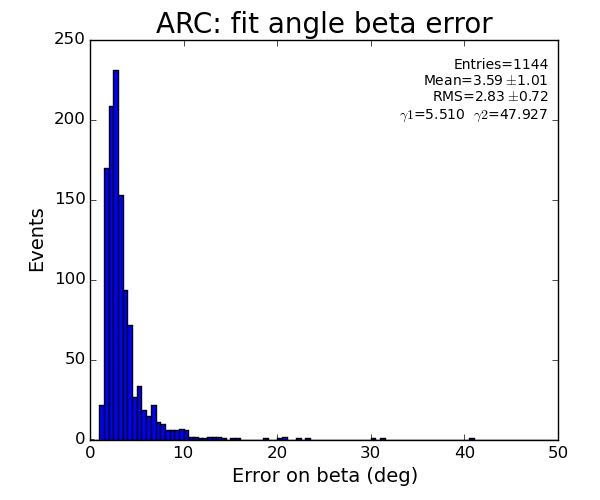
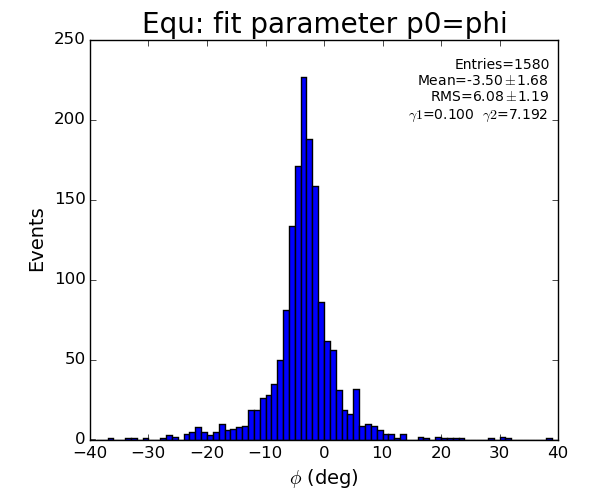
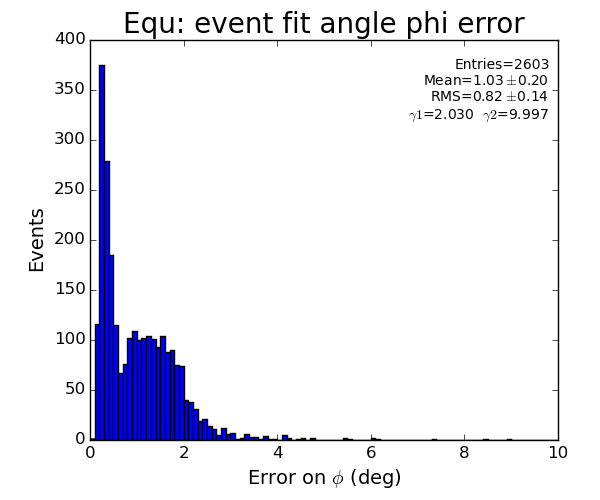
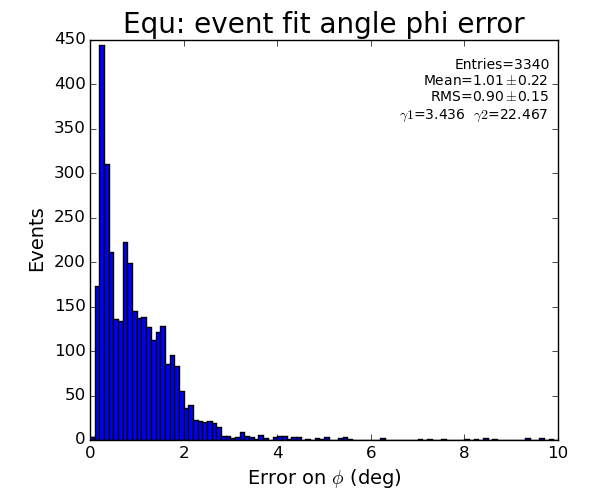
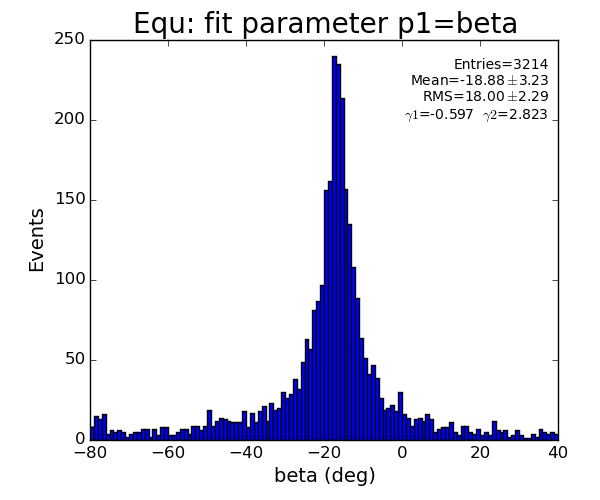
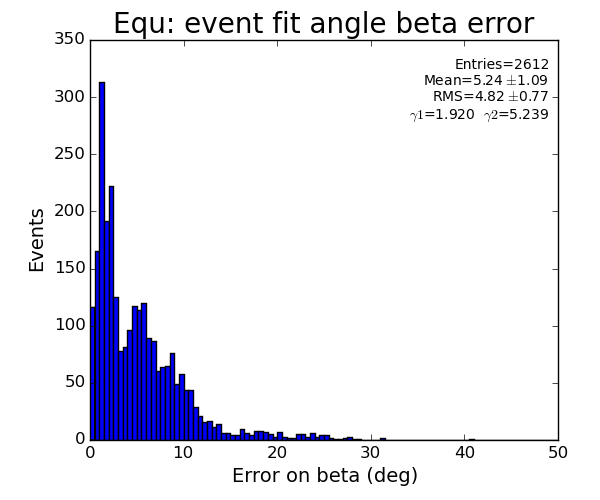
Fit results
ARC
pre-fit selection
def peakIsSelectedArc(pk) :
"""Apply peak selection criteria to each peak from file
"""
if sp.USE_PEAK_SELECTION :
if pk.atot<1500 : return False
if pk.npix>200 : return False
if pk.r<435 : return False
if pk.r>443 : return False
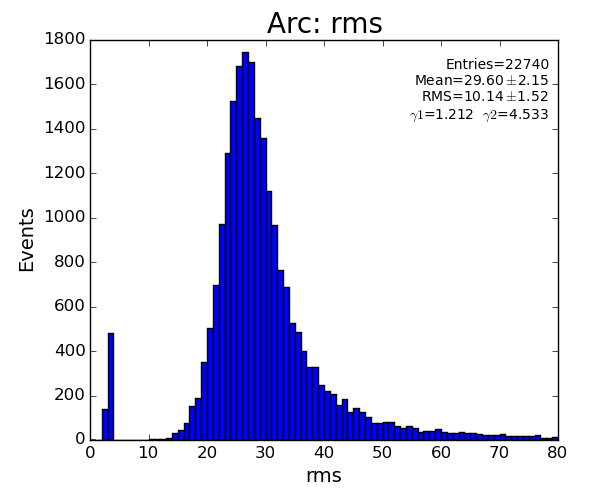
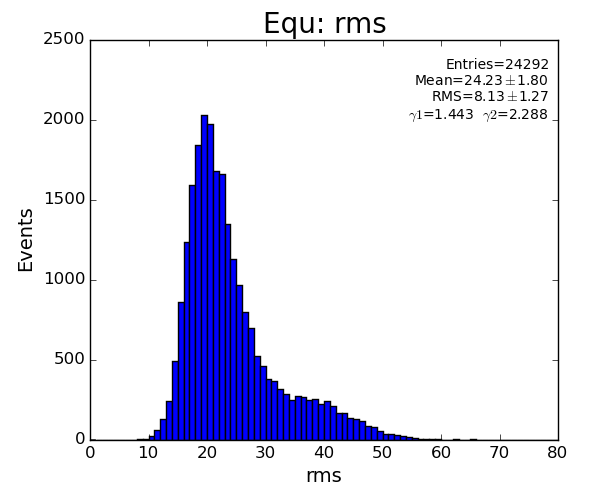
if pk.rms<10 : return False
if pk.rms>80 : return False
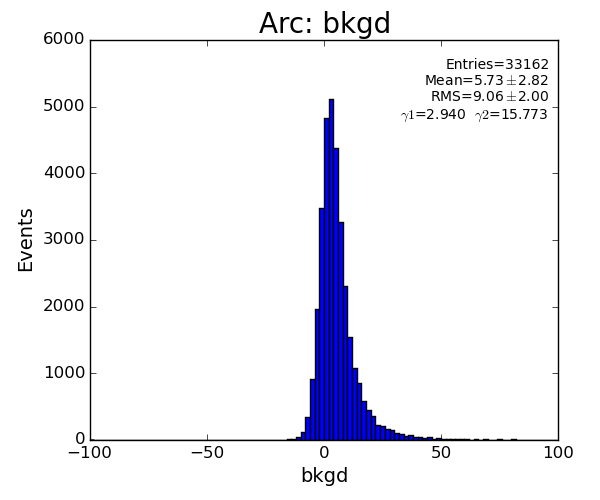
if math.fabs(pk.bkgd)>25 : return False
return True
plots for pfv2/3/4r1
================
EQU
pre-fit selection
def peakIsSelectedEqu(pk) :
"""Apply peak selection criteria to each peak from file
"""
if sp.USE_PEAK_SELECTION :
if pk.atot<1500 : return False
if pk.npix>200 : return False
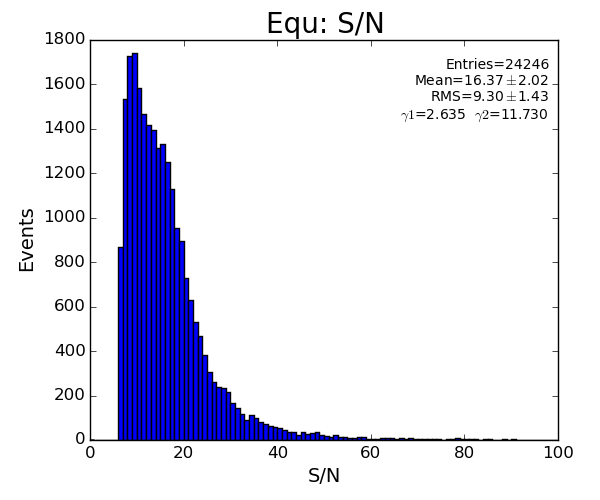
if pk.rms>80 : return False
if math.fabs(pk.bkgd)>20 : return False
if pk.r<100 : return False
if pk.r>454 : return False
return True
plots for pfv2/3/4r1
================
References
- Masks, Background subtraction
- PSAS-147 pfv3r1 - is upgraded
- Hit and Peak Finding Algorithms - S/N statistical significance is improved by increasing number of pixels
Overview
Content Tools