Psocake Tutorial
Sections in this tutorial
Psocake is a graphical user interface (GUI) for looking at detector images at LCLS. By the end of this tutorial, you will be able to generate mask, find peaks in your images and index crystal diffraction patterns.
- Starting psocake
- Mask making
- Peak finding
- Jumping to interesting images based on the number of peaks
Indexing crystals
Starting psocake
Type "psocake" on your terminal to open up the GUI:
$ psocake
1) There are four parameters required to uniquely identify an image at LCLS. Type the experiment name, run number, detector name, and event number in the Experiment Parameters panel.
For this tutorial, we will look at experiment cxitut13, run 10, detector DscCsPad, event 11.
####################### # Available detectors: ['DscCsPad'] #######################
2) You can specify the experiment parameters as command line arguments in psocake using the psana-style experiment run string:
$ psocake exp=cxitut13:run=10 -d DscCsPad -n 11
3) You can also use the -e and -r arguments for the experiment and the run number:
$ psocake -e cxitut13 -r 10 -d DscCsPad -n 11
Don’t worry if you don’t remember these arguments. For argument options, use --help:
$ psocake --help
Mask making
In this example, let's mask out pixels that should not be used for analysis, mask out the jet streak at the centre of the detector, and mask out the water ring.
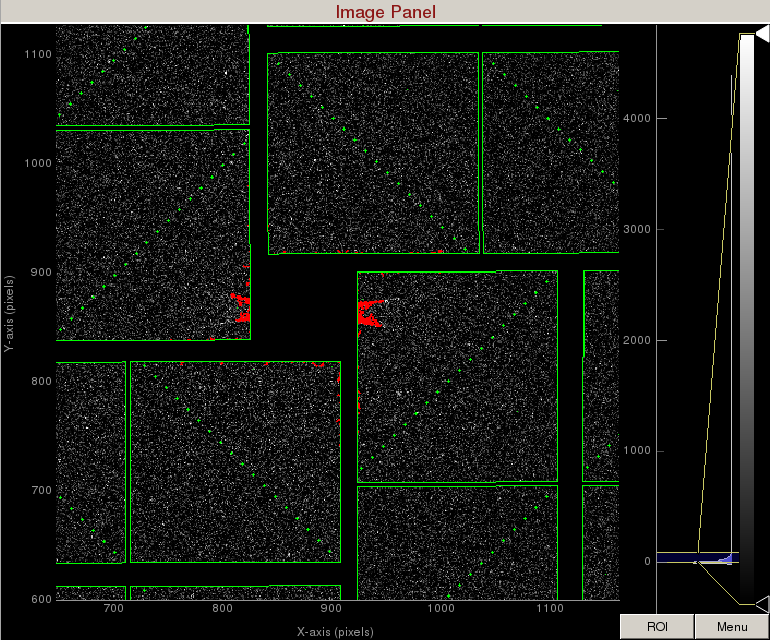
1) On the mask panel, click on "Use psana mask". This will mask out the following pixels that should not be used for analysis; calib, status, edge, central, unbonded pixels, unbonded pixel neighbor pixels. These pixels are shown as green on the image panel.
2) On the mask panel, click on "Use streak mask". This will mask out strong intensities originating from the edges of the central asics.
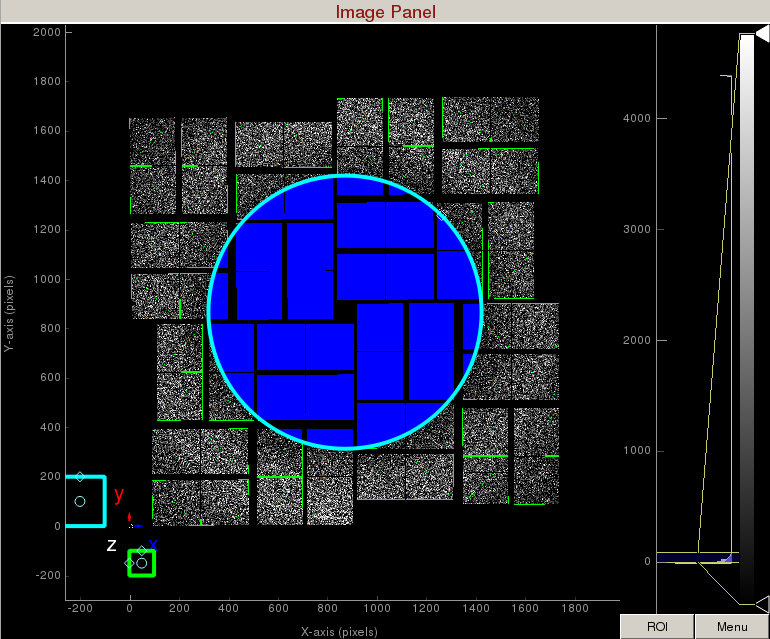
3) To make a donut mask over the water ring, click on "Use user-defined mask". This will bring up a cyan circle and cyan square mask generator.
Select "Toggle" in Masking mode. Move the cyan circle to the centre of the detector by dragging the circle. Resize the cyan circle by dragging the diamond on the perimeter. Once you are happy with the position, click "mask circular ROI" button on the mask panel.
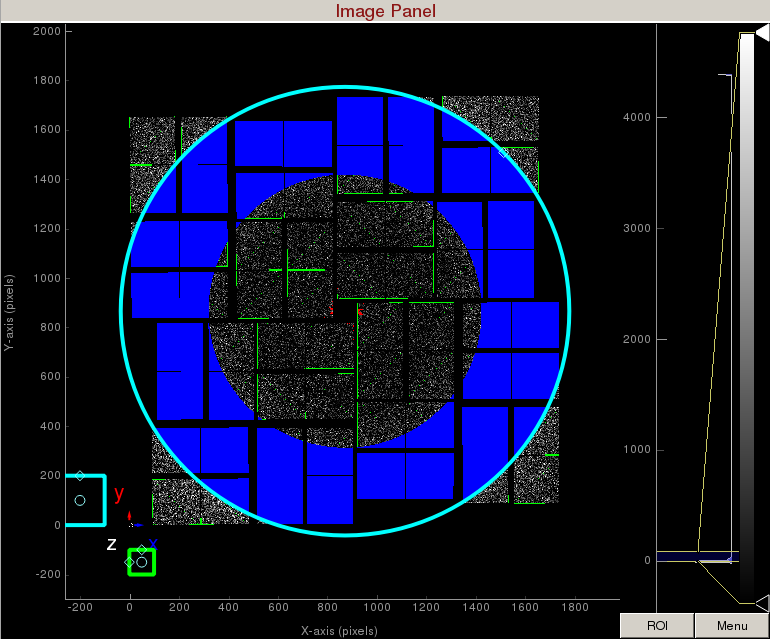
Increase the cyan circle again by dragging the diamond on the perimeter. Click "mask circular ROI" button on the mask panel. Because we are in the "toggle" mode, the previous mask gets toggled and disappears. The area that does not overlap with the previous mask get masked out.
To save the user-defined mask, click on "Save user-defined mask" on the mask panel which will save the mask in the scratch folder. For this example, the mask will be saved here:
/reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/mask.npy (3D ndarray)
/reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/mask.txt (2D text)
You can load the user-defined mask using the "Load user-defined mask" button and selecting mask.npy.
Peak finding
- Set npix_min: 2
- Set npix_max: 5
- radius: 2
Hover the mouse pointer over the Bragg peaks to study the intensities. The sum of the Bragg peak pixels are above 400 ADUs. Set the following values:
- Set atot_thr: 400
- thr_low: 160
- thr_high: 160
- son_min: 4
The output directory on the peak finder panel should already be automatically set to: /reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake
- CPUs: 3
- Number of events to process: 30
Jumping to interesting images based on the number of peaks
In the small data panel, type the CXIDB filename:
- filename: /reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/cxitut13_0010.cxi
- metric_dataset: /entry_1/result_1/nPeaksAll
Indexing crystals
Let's try to index the diffraction pattern at event 11.
- In the experiment parameters panel, set Event Number to 11.
In the indexing panel, tick "Indexing on". Indexed peaks will be marked with magenta circles. These triple rings represent the integration radius.
Hopefully, you have indexed this diffraction pattern. Notice that the unitcell parameters are a bit off compared to what is expected. Let's load a CrystFEL unitcell file to help the indexer along.
- In the indexing panel, set the PDB field to: /reg/d/psdm/cxi/cxitut13/scratch/psocake/lys.cell
CrystFEL unit cell file version 1.0 lattice_type = tetragonal centering = P unique_axis = c a = 77.05 A b = 77.05 A c = 37.21 A al = 90 deg be = 90 deg ga = 90 deg
- In the diffraction geometry panel, set "Detector distance" to 156mm. Try incrementing this distance in 1mm step till the unit cell parameters are as close as possible to lys.cell. The optimum detector distance is around 158mm.
- Try drawing a resolution ring on top of the water ring by using “Resolution (pixels)” field. If your detector distance is correct, the crystallographic resolution ring should display 3.4A.
Future plans
- More mask shapes
- More integration with logbook
- Extend the crystallography analysis to XPP and MFX beamlines
- cctbx-xfel support