Page History
...
| Code Block | ||
|---|---|---|
| ||
Citation for psocake (and other psana-based programs):
@Article{Thayer2017,
author="Thayer, J. and Damiani, D. and Ford, C. and Dubrovin, M. and Gaponenko, I. and O'Grady, C. P. and Kroeger, W. and Pines, J. and Lane, T. J. and Salnikov, A. and Schneider, D. and Tookey, T. and Weaver, M. and Yoon, C. H. and Perazzo, A.",
title="Data systems for the Linac coherent light source",
journal="Advanced Structural and Chemical Imaging",
year="2017", month="Jan", day="14", volume="3", number="1", pages="3", issn="2198-0926",
doi="10.1186/s40679-016-0037-7", url="https://doi.org/10.1186/s40679-016-0037-7"}
@article{Damiani:zw5004,
author = "Damiani, D. and Dubrovin, M. and Gaponenko, I. and Kroeger, W. and Lane, T. J. and Mitra, A.
and O'Grady, C. P. and Salnikov, A. and Sanchez-Gonzalez, A. and Schneider, D. and Yoon, C. H.",
title = "{Linac Coherent Light Source data analysis using {it psana}}",
journal = "Journal of Applied Crystallography",
year = "2016", volume = "49", number = "2", pages = "672--679", month = "Apr",
doi = {10.1107/S1600576716004349}, url = {http://dx.doi.org/10.1107/S1600576716004349}, } |
Please connect to psana machines via NoMachine for less lag in graphics. Instructions here: Remote Visualization
Starting psocake in SFX mode
...
| Code Block | ||
|---|---|---|
| ||
# PSOCAKE2 (Run 17 or older)
function psocake2 ()
{
echo "Activating psocake py2"
source /reg/g/psdm/etc/psconda.sh
# CrystFEL compatible version for psocake2 -py2
source /reg/g/cfel/crystfel/crystfel-0.8.0/setup-sh # CrystFEL compatible version for psocake2
}
# PSOCAKE3 (Run 18 or newer)
function psocake3 ()
{
echo source"Activating psocake py3"
source /reg/g/psdm/etc/psconda.sh -py3
# Crystfel compatible version for psocake3
source /reg/g/cfel/crystfel/crystfel-0.9.1/setup-sh # Crystfel compatible version for psocake3
}
# CCP4 (version may change)
source /reg/common/package/ccp4-7.1/bin/ccp4.setup-sh
# XDS
export PATH=/reg/common/package/XDS-INTEL64_Linux_x86_64:$PATH |
Then source ~/.bashrc
For LCLS Run 18 or newer runs, call the psocake3 function by typing psocake3 in the terminal.
For older LCLS runs, use psocake2. .
| Code Block | ||
|---|---|---|
| ||
$ source ~/.bashrc
$ psocake3
Activating psocake py3 |
For this tutorial, we will look at experiment cxic00318, run 124, detector jungfrau4M, event 33.
...
Mask making
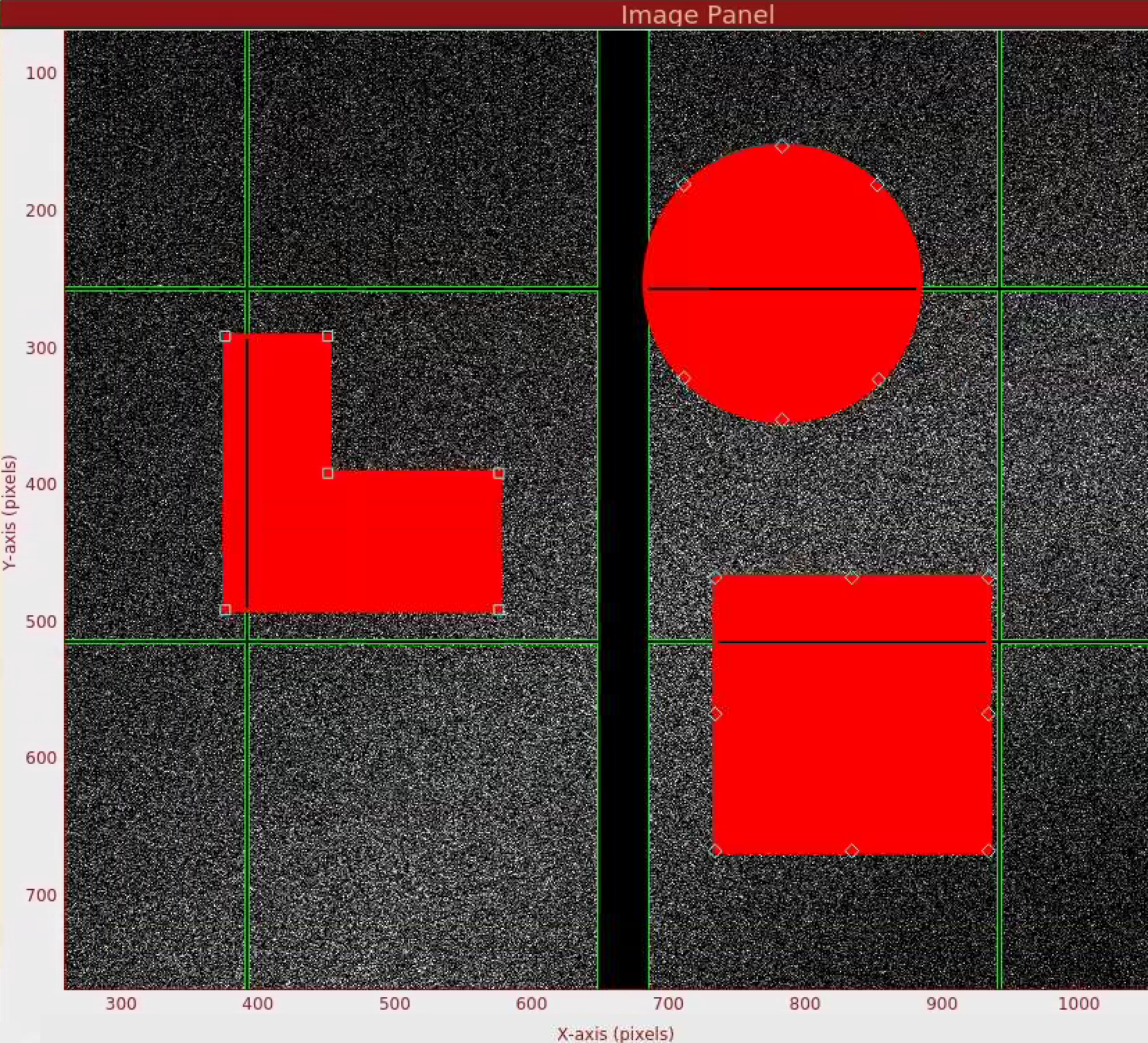
In this section, let's you will learn how to mask out pixels that should not be used for analysis (such as dead pixels and shadows), mask out the jet streak at the centre of the detector, and mask out the water ring (just for fun!). Masked out regions are ignored during peak finding and during Bragg spot integration.
Note: the Image Panel must be in the default "greyscale" colormap for the mask colors to display properly.
...
1) In the mask panel, "Use psana mask" checkbox is turned on by default.
This will mask out the following pixels that should not be used for analysis; calib, status, edge, central, unbonded pixels, unbonded pixel neighbor pixels. Hovering over the checkboxes shows what these options mean. These masked pixels are shown as green on the image panel. You can selectively turn these on and off. Red circle indicates the detector centre.
2) To mask out jet streak near the centre of the detector, click on "Use streak mask".
This will mask out strong intensities originating from the liquid jet. The streak mask is dynamic, i.e. recalculated for each image.
3) Let's look at using the mask widgets.
In the Mask Panel, turn on "Use user-defined mask". Select "Mask" in Masking mode. This will bring up a red rectangular, circle, and polygon and squaremask widgets on the left of the detector image. You can drag a mask widget onto the detector by clicking and holding the mouse button inside a widget. There are diamond handles on these widgets which you can drag to resize. For the polygon widget, you can create additional handles (or corners) by a single click on the edge of the widget. Once positioned, use the "Stamp rectangular mask", "Stamp circular mask", and "Stamp polygon mask" buttons to create the mask. In this "Mask" mode, you can click on single pixels on the detector to make out single pixels. "Mask outside histogram" button will mask out pixels that are below the histogram low threshold and above the histogram high threshold.
To save the user-defined mask, click on "Save static mask" button on the mask panel which will save the mask in your designated psocake run folder. This will combine user-defined mask (red) and the psana mask (green) into a single mask. You should see the following message in the terminal:
...
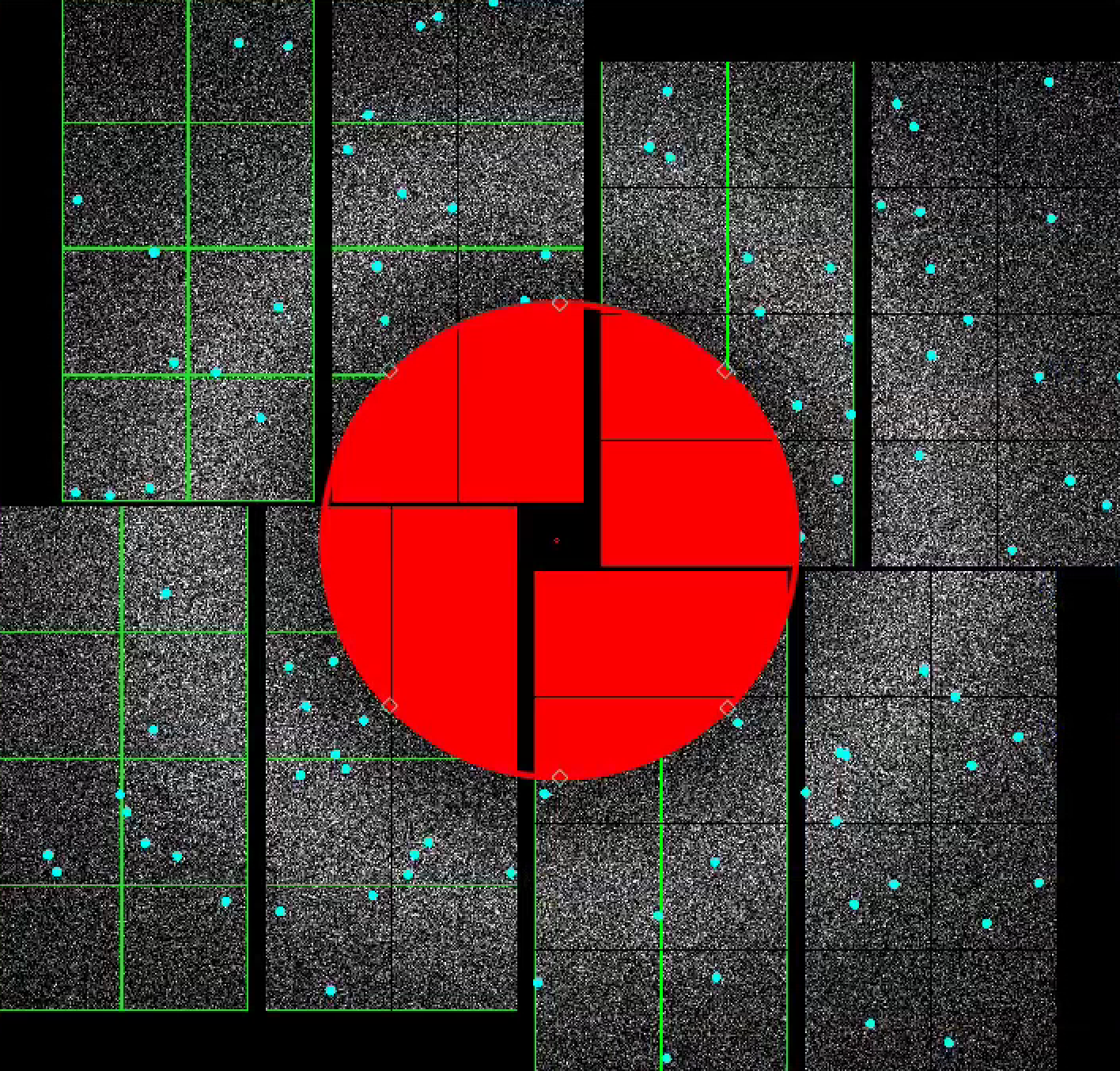
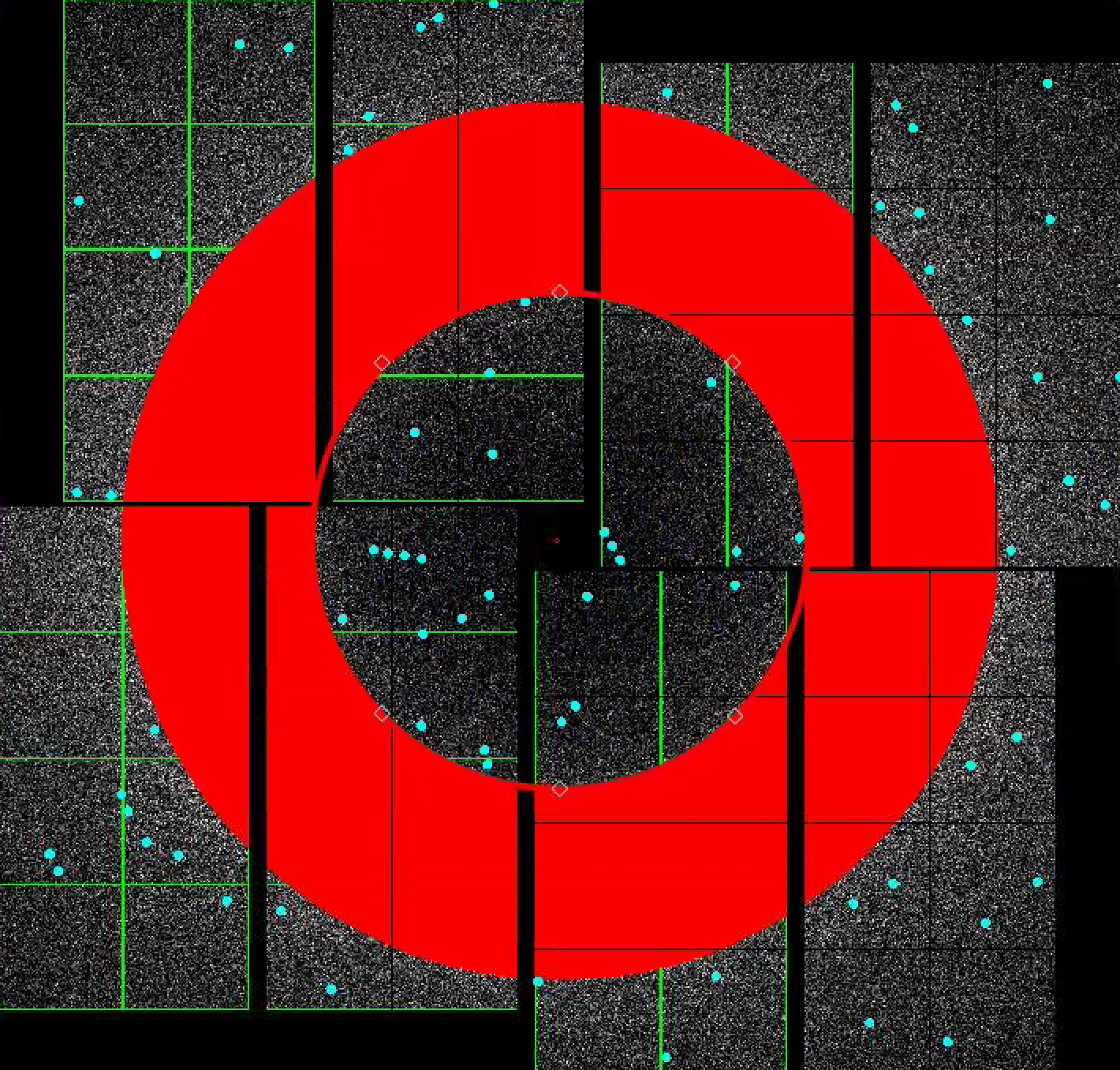
4) To make a donut mask over the water ring, turn turn on "Use user-defined mask". Select "Toggle" in Masking mode.
Move the red circle widget to the centre of the detector by dragging it. Resize it by dragging the diamond handle on the perimeter. Once you are happy with the area to be masked out, click "Stamp circular mask" button on the mask panel.
Increase the size of the red circle widget again by dragging the diamond handle on the perimeter. Click "Stamp circular mask" button on the mask panel. Because we are in the "toggle" mode, previously mask pixels inside our widget is unmasked, i.e. gets toggled. The area that does not overlap with the previous mask get masked out.
Peak finding
In this section, we will find peaks on the detector image.
...
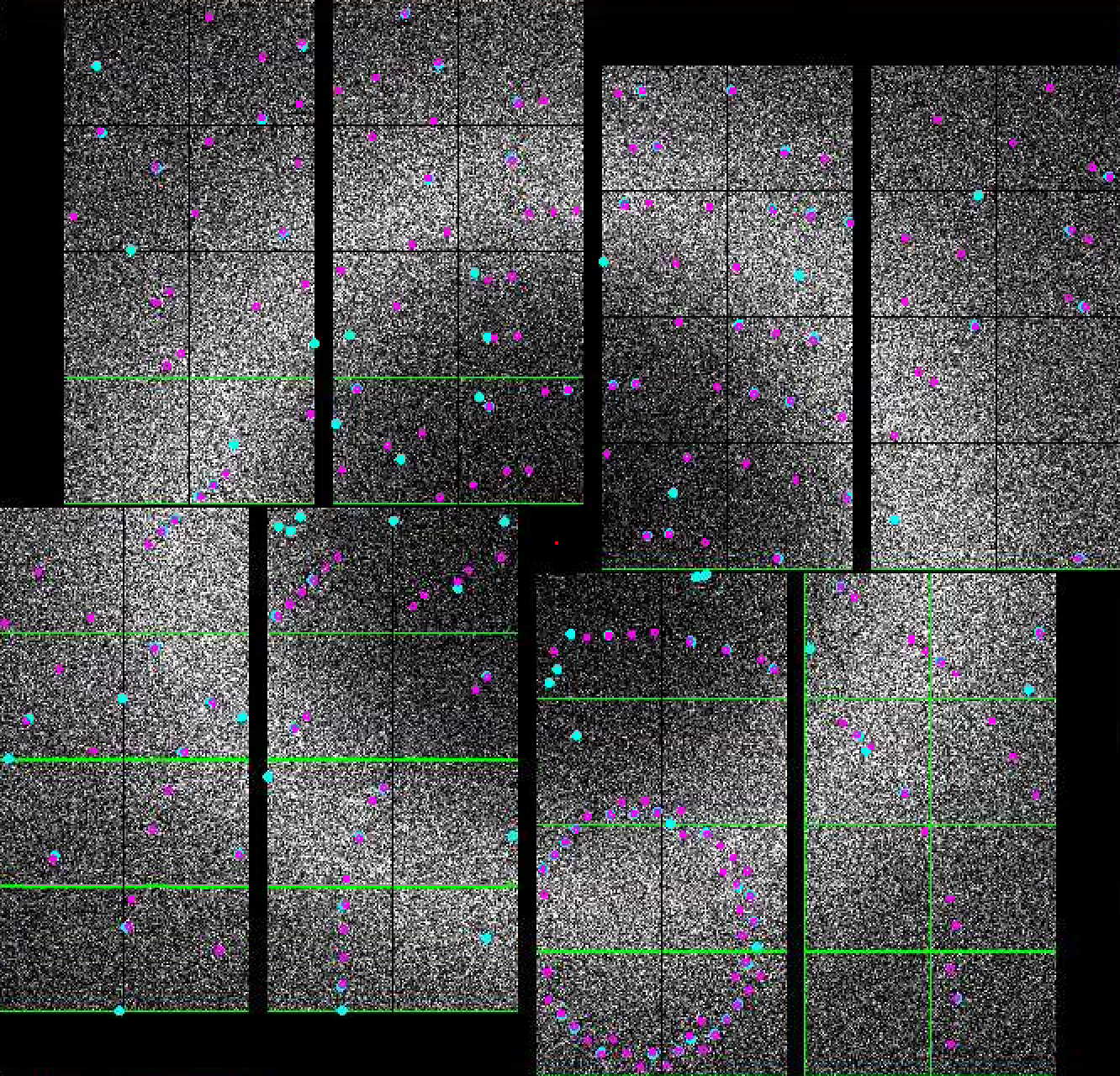
Default setting in the Peak Finder panel is to "Show peaks found". You should notice peaks being highlighted in the Image panel. If you are happy with the peaks found, you are done. Otherwise, you can interactively tune peak finding parameters by expanding the "Parameters" field. Details of the peak finding algorithm is given here: Hit and Peak Finding Algorithms.
...
First things first, crystal indexing requires an accurate detector geometry. Latest CXI geometry files can be found here: Geometry history
Get in touch with the beamline POC if the geometry is inaccurate.
- In the indexing panel, tick "Indexing on". If indexing succeeds, the predicted peaks will be marked with
- In the diffraction geometry panel, set "Detector distance" to 145mm.
- In the indexing panel, tick "Indexing on". If indexing succeeds, the integrated (predicted) peaks will be marked with magenta circles. These triple rings represent the integration radii. A large central magenta triangle means means wait. If If indexing fails, you will see a a large central magenta X.
- If you see magenta circles and magenta unitcell appear, congratulations! You have indexed your first pattern using psocake.
- Try incrementing this distance in 1mm step till the unit cell parameters are as close as possible to lys.cell. The optimum detector distance is around 145mm.
Every time the "Detector distance" value is changed by the user, psocake converts the psana geometry (in /reg/d/psdm/cxi/cxic00318/calib/Jungfrau\:\:CalibV1/CxiDs1.0\:Jungfrau.0/geometry/123-end.data) to a CrystFEL geom file (in /reg/d/psdm/cxi/cxic00318/scratch/<username>/psocake/<runNumber>/.temp.geom).
...
- Run(s): 124
- Queue: psanaq
- CPUs: 24
- Keep CXI images: On
Indexing will take some time to complete. If successful, you should see a stream file in: /reg/d/psdm/cxi/cxic00318/scratch/<username>/psocake/r0124/cxic00318_0124.stream
You can check the status of your indexing job here: /reg/d/psdm/cxi/cxic00318/scratch/<username>/psocake/r0124/status_index.txt
Psocake saves the detector images of only the hits in the .cxi file. It is likely that you may want to reindex these files to optimize the indexing rate. If you anticipate that you have finalized the indexing parameters, set 'Keep CXI images' to Off. It will delete the detector images in your .cxi file which will free up your precious disk space for doing other things.
As with peak finding, you can launch indexing jobs on multiple runs by specifying runs in the Run(s) field.
Indexing multiple lattices
Save publication images
You can save publication quality images by clicking "Save evt" button near the lower bottom right of the GUI.
Image: psocake_cxic00318_124_jungfrau4M_30_1597321015_275824216_83040_img.png
Peaks found: psocake_cxic00318_124_jungfrau4M_30_1597321015_275824216_83040_pks.png
Indexed peaks: psocake_cxic00318_124_jungfrau4M_30_1597321015_275824216_83040_idx.png
CrystFEL can index multiple lattices per image. Set "Extra CrystFEL parameters" to --multi,--no-check-peaks. This will enable "subtract and retry" method.
Indexing pump-probe experiments
In a pump probe experiment, it is sometimes desirable to index only certain events, e.g index only the crystals where the optical pump laser was on. This information is recorded in the EVR which psocake saves in the .cxi file (dataset name: "/LCLS/detector_1/evr1").
So if you want to index only the hits with laser on (say EVR1: 182), then type the following in the "Index condition" field:
182 in #evr1#
Psocake will also accept combinations using AND/OR:
182 in #evr1# and 173 in #evr1#
You can attach a tag to the stream filename by using the "Tag" field in the Indexing panel, e.g. evr182 would produce a new stream file cxitut13_0010_evr182.stream.
Detector centering
Let's check whether your detector is well centered with respect to your beam. You want the centre to be as accurate as possible (at least to a pixel accuracy) for optimal indexing rates.
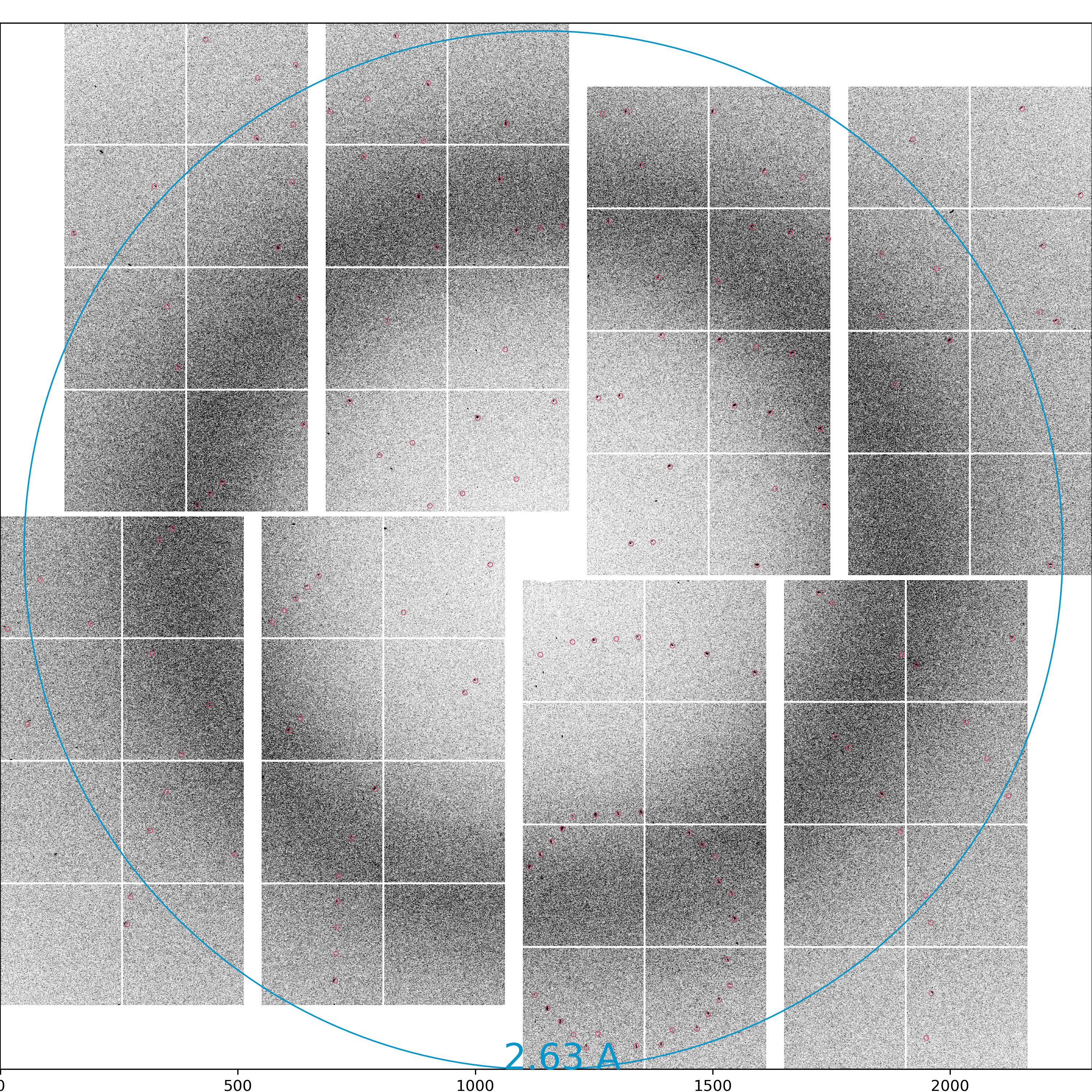
Load the powder rings generated by clicking the "Load image" button in the Image Control panel. Open "cxic00318_0124_maxHits.npy". Adjust the intensity as necessary.
Draw resolution rings by ticking "Resolution rings" in the Diffraction Geometry panel. You can change the ring resolution by typing number in "Resolution (pixels)". Type 165 and see whether your powder rings overlap with the resolution rings. If they do, then the detector is centered. If not, then you can click on the "Deploy automatically centred geometry" to recenter your detector. If you are unhappy with the results, you can use "Deploy manually centred geometry" which will shift the detector centre to the centre of the green ROI circle.
Since we are at run124, the newly deployed geometry file is named 124-end.data. If there already exists a geometry file with the same name, it will be backed up as 124-end.data-<timeModified>
Detector geometry optimization
Past geometry files can be found here: Geometry history
It is often the case at the beginning of a beamtime that the detector distance to the interaction point (coffset) is not precisely known (we are talking about sub-millimetre precision), and we can use the Diffraction Geometry panel to find the optimal distance.
Post-processing
| Code Block | ||
|---|---|---|
| ||
# Phenix (version number may change) source /reg/common/package/phenix/phenix-1.10.1-2155/phenix_env.sh # CCP4 (version number may change) source /reg/common/package/ccp4/ccp4-7.0/bin/ccp4.setup-sh # XDS is available here /reg/common/package/XDS-INTEL64_Linux_x86_64/ |
For viewing the electron density, use coot contained inside phenix.
Beam Parameters for Publications
| Code Block | ||||
|---|---|---|---|---|
| ||||
ebeam = ebeamDet.get(evt) pulseEnergy = ebeam.ebeamL3Energy() # MeV es = psana.DataSource.env().epicsStore() calculatedNumberOfPhotons = get_es_value(es, 'SIOC:SYS0:ML00:AO580', NoneCheck=False, exceptReturn=0) * 1e12 # photons |
Bug/Comments:
Please send bug reports/comments:
Tiny url for this tutorial: http://tinyurl.com/zj4m23n