"Data analysis == Piece of cake"
Sections in this tutorial
It is always a good idea for the people doing analysis to be able to look at their detector images and probe intensity values. Given that a typical LCLS experiment has millions of snapshots to choose from, it is also critical that you can quickly select images of interest and set regions of interest using masks. By the end of this tutorial, you will be able to browse images, jump to images of interest, generate masks, and find hits.
Citation for psocake (and other psana-based programs):
@article{Damiani:zw5004,
author = "Damiani, D. and Dubrovin, M. and Gaponenko, I. and Kroeger, W. and Lane, T. J. and Mitra, A.
and O'Grady, C. P. and Salnikov, A. and Sanchez-Gonzalez, A. and Schneider, D. and Yoon, C. H.",
title = "{Linac Coherent Light Source data analysis using {it psana}}",
journal = "Journal of Applied Crystallography",
year = "2016", volume = "49", number = "2", pages = "672--679", month = "Apr",
doi = {10.1107/S1600576716004349}, url = {http://dx.doi.org/10.1107/S1600576716004349}, }
Starting psocake in SPI mode
Type "psocake" on your terminal to open up the GUI. For single particle imaging, we will need to open it in spi mode (-m):
$ psocake -m spi
1) There are four parameters required to uniquely identify an image at LCLS. Type the experiment name, run number, detector name, and event number in the Experiment Parameters panel.
For this tutorial, we will look at experiment cxitut13, run 10, detector DscCsPad, event 11.
#######################################
# Available area detectors:
# ('CxiDs1.0:Cspad.0', 'DscCsPad', '')
#######################################
2) You can specify the experiment parameters as command line arguments in psocake using the psana-style experiment run string:
$ psocake exp=cxitut13:run=10 -d DscCsPad -n 11 -m spi
3) You can also use the -e and -r arguments for the experiment and the run number:
$ psocake -e cxitut13 -r 10 -d DscCsPad -n 11 -m spi
To check psocake version:
$ psocake --version
Don’t worry if you don’t remember these arguments. For argument options, use --help:
$ psocake --help
Mask making
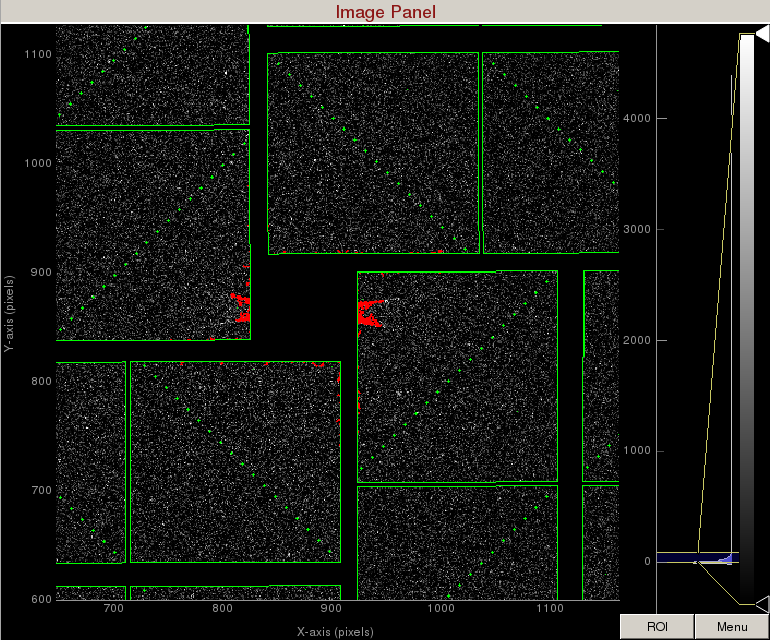
In this example, let's mask out pixels that should not be used for analysis, mask out the jet streak at the centre of the detector, and mask out the water ring.
Note: the Image Panel must be in the default "greyscale" colormap for the mask colors to display properly.
1) On the mask panel, click on "Use psana mask". This will mask out the following pixels that should not be used for analysis; calib, status, edge, central, unbonded pixels, unbonded pixel neighbor pixels. These pixels are shown as green on the image panel.
2) On the mask panel, click on "Use streak mask". This will mask out strong intensities originating from the edges of the central asics.
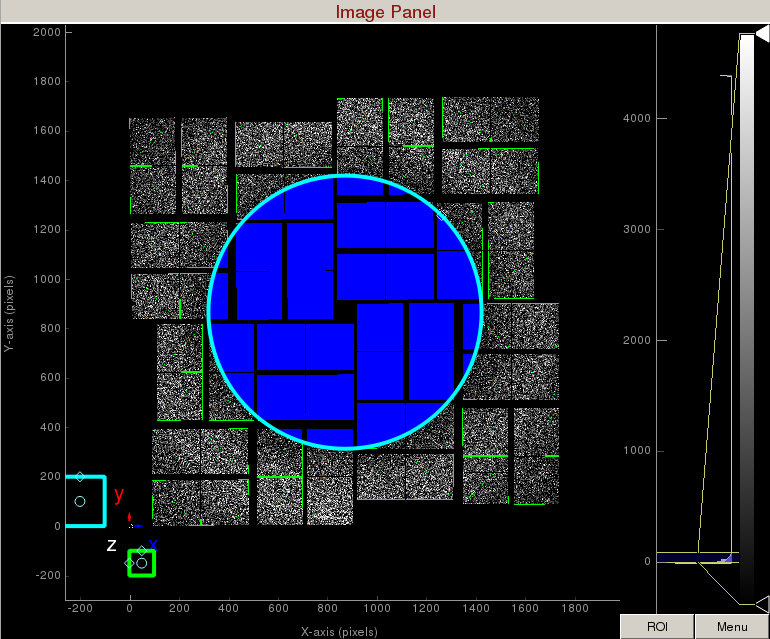
3) To make a donut mask over the water ring, click on "Use user-defined mask". This will bring up a cyan circle and cyan square mask generator.
Select "Toggle" in Masking mode. Move the cyan circle to the centre of the detector by dragging the circle. Resize the cyan circle by dragging the diamond on the perimeter. Once you are happy with the position, click "mask circular ROI" button on the mask panel.
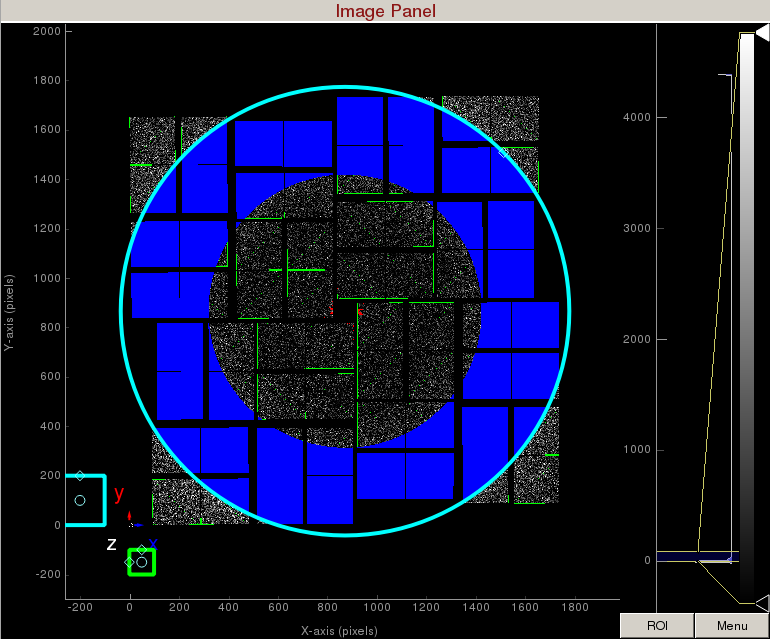
Increase the cyan circle again by dragging the diamond on the perimeter. Click "mask circular ROI" button on the mask panel. Because we are in the "toggle" mode, the previous mask gets toggled and disappears. The area that does not overlap with the previous mask get masked out.
To save the user-defined mask, click on "Save static mask" on the mask panel which will save the mask in the scratch folder. For this example, the mask will be saved here:
/reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/mask.npy (3D ndarray)
/reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/mask.txt (2D text)
You can load the user-defined mask using the "Load mask" button and selecting mask.npy.
Hit finding
- Set ADUs per photon: 200
- Run(s): 10
- CPUs: 2
- Number of events to process: 30
Jumping to interesting images based on hit finder
In the small data panel, you should already see the CXIDB filename and dataset displayed:
- filename: /reg/d/psdm/cxi/cxitut13/scratch/<username>/psocake/r0010/cxitut13_0010.cxi
- dataset: /entry_1/result_1/nHitsAll
Saving hits in cxidb format
Please send bug reports/comments: