Content
Phi-beta fit functions
Two solutions of quadratic equation in ARC region
- POS: y = -B + sqrt(B*B-C)
- NEG: y = -B - sqrt(B*B-C)
Arc region
pyimgalgos/src/FiberAngles.py
y = funcy_l1_v1(x, phi_deg, bet_deg, DoR=390/913.27, sgnrt=1.) # or ... sgnrt=-1.)
wide range of x:
narrow range of x:
- For POS root: beta=-13.3 gives minimal y.
- If two measured peaks are located below this curve then B*B-C < 0, fit returns limit value of beta or not converging.
- Two solutions for beta -17 and -9 - x and y are the same.
- NEG-ative root gives wrong curvature
Equatorial region
pyimgalgos/src/FiberAngles.py
y = funcy_l0(x, phi_deg, bet_deg)
Problem with two roots is resolved for EQU region; sign of the root is selected the same as sign of parameter B;
- For |beta|>~48 solution does not exist for entire range of x
Fit in the processing script
Example of how to apply fit to data using absolute errors on peak position, get fit parameters with errors, estimate fit quality.
cxif5315/proc-cxif5315-r0169-peaks-from-file-v4.py, v5, v6
from scipy.optimize import curve_fit
from scipy.stats import chi2
xn = np.array([pk.x/L for pk in sp.lst_equ_evt_peaks], dtype=np.double)
yn = np.array([pk.y/L for pk in sp.lst_equ_evt_peaks], dtype=np.double)
en = np.array([pk.csigma*sp.PIXEL_SIZE/L for pk in sp.lst_equ_evt_peaks], dtype=np.double)
#en = np.array([max(pk.rsigma,pk.csigma)*sp.PIXEL_SIZE/L for pk in sp.lst_equ_evt_peaks], dtype=np.double)
p0 = [-3.2,-16.0] # phi, beta angle central values
popt, pcov = curve_fit(funcy, xn, yn, p0, en, absolute_sigma=True)
sp.fib_chi2 = np.sum(((funcy(xn, *popt) - yn) / en)**2)
sp.fib_ndof = len(xn) - 1
sp.fib_prob = chi2.sf(sp.fib_chi2, sp.fib_ndof, loc=0, scale=1)
Phi-beta fit results and quality
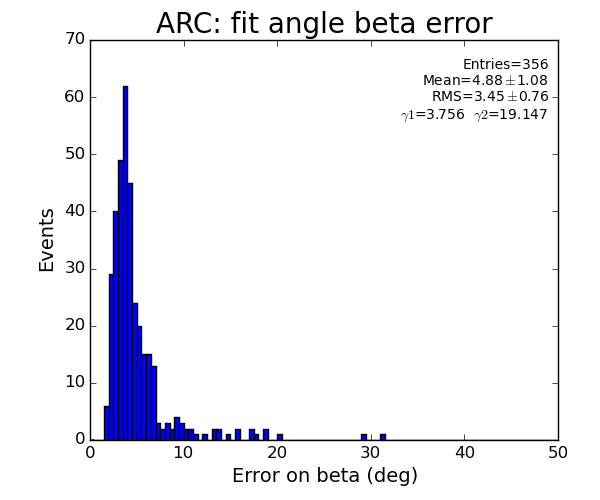
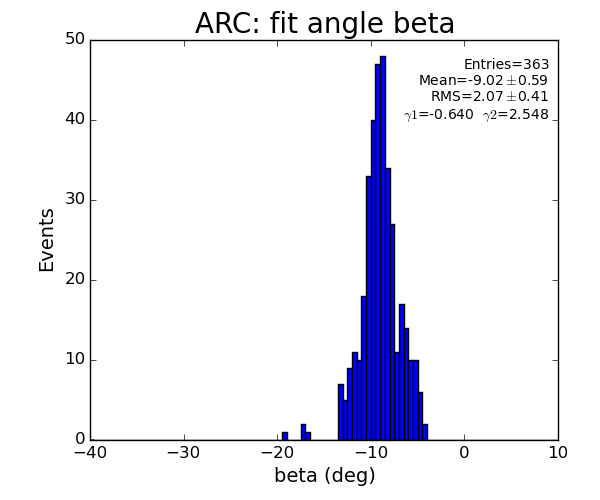
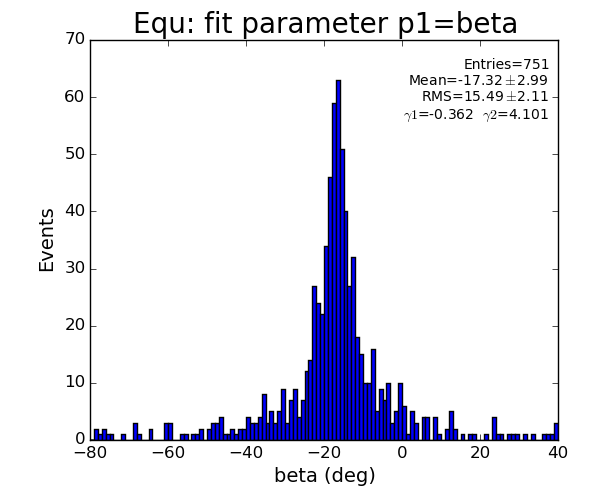
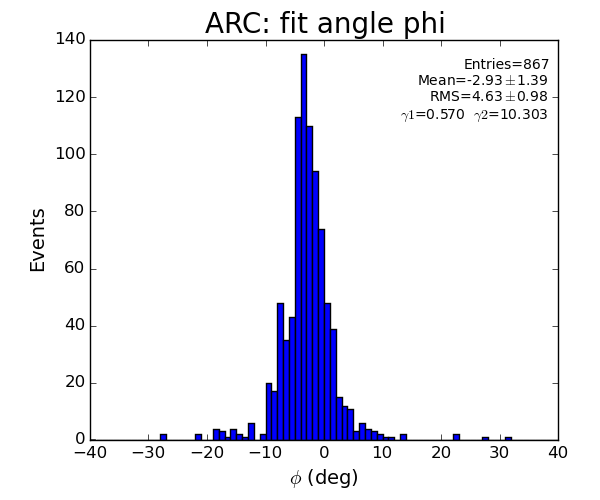
Data: peak list selected by Meng
- exp=cxif5315:run=169
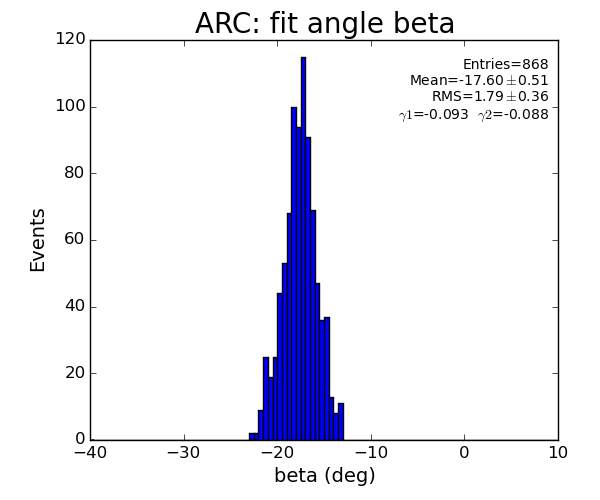
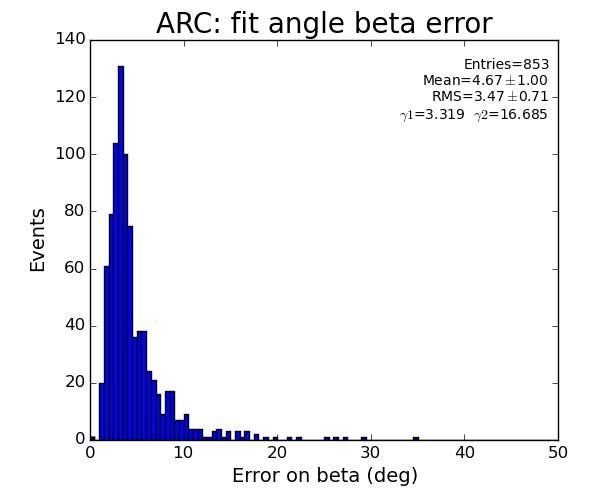
- Distribution of parameters for peaks selected by Meng:
EQU region
ARC region: 1-st solution for beta (positive root)
ARC region: 2-nd solution for beta (positive root)
For 2-peak events fit does not have any freedom - curve can exactly be fitted to 3 points (including origin); chi2=0 and probability is always 1.
For >2-peak events chi2 and probability show some distribution.
Data: peak list after pfv2r1
EQU region
ARC region
Peak list with phi-beta fit results
After phi-beta fit peaks are saved in the file with name like
result/peak-fit-cxif5315-r0169-2016-04-04T16:16:38.txt
Comparing to regular peak parameters this file has extended data for each peak by
... fit-phi fit-beta phi-err beta-err fit-chi2 ndof fit-prob ... 1.30 -42.98 3.800454 10.236941 0.000000 1 1.000000 ... -4.75 -23.85 0.265462 1.212093 1.734365 3 0.629321
To load this file use helper classes pyimgalgos.TDFileContainer with pyimgalgos.TDPeakRecord
References