COLTRIMS (Cold Target Recoil Ion Momentum Spectroscopy)
Walter, Peter <pwalter@slac.stanford.edu> Mon 6/3, 4:29 PM O'Grady, Paul Christopher; Thayer, Jana B.; Weninger, Clemens; Dubrovin, Mikhail, DAQ Control Dear all, Runs with coincidence measurements are 85, 86, and 88-93 from amox27716 A good run for covariance would be run 100. And here is the code Xiang wrote with the help from Clemens and I. https://github.com/xianglgithub/Coinc-LCLS |
on 2019-06-25 runs 91, 92, 93, 100 requested to restore from tape
/reg/d/psdm/AMO/amox27716/xtc/
exp=amox27716:run=100
event_keys -d exp=amox27716:run=100
Event# 9 EventKey(type=psana.EvrData.DataV4, src='DetInfo(NoDetector.0:Evr.0)') EventKey(type=psana.Acqiris.DataDescV1, src='DetInfo(AmoEndstation.0:Acqiris.1)', alias='ACQ1') EventKey(type=psana.Acqiris.DataDescV1, src='DetInfo(AmoEndstation.0:Acqiris.2)', alias='ACQ2') EventKey(type=psana.Camera.FrameV1, src='DetInfo(AmoEndstation.0:Opal1000.3)', alias='OPAL3') EventKey(type=psana.Camera.FrameV1, src='DetInfo(XrayTransportDiagnostic.0:Opal1000.0)', alias='xtcav') EventKey(type=psana.Bld.BldDataEBeamV7, src='BldInfo(EBeam)') |
For exp=amox27716:run=100 in LCLS1 environment
hex/examples/ex_acqiris_quad_amox27716-0100.py
dsname = 'exp=amox27716:run=100'
src1 = 'AmoEndstation.0:Acqiris.1' # 'ACQ1'
src2 = 'AmoEndstation.0:Acqiris.2' # 'ACQ2'
Acqiris.1:
nbrChannels: 8, H-nbrSamples: 44000, H-sampInterval: 2.5e-10
chan: 0, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000015, V-offset: 0.000000, H-pos[seg=0]: -7.8262e-11
chan: 1, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000076, V-offset: -2.000000, H-pos[seg=0]: -7.8262e-11
chan: 2, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
chan: 3, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
chan: 4, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
chan: 5, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
chan: 6, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
chan: 7, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 0, V-slope: 0.000031, V-offset: 0.800000, H-pos[seg=0]: -7.8262e-11
wf.shape: (8, 44000) wt.shape: (8, 44000)
Acqiris.2:
nbrChannels: 2, H-nbrSamples: 44000, H-sampInterval: 2.5e-10
chan: 0, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 2, V-slope: 0.000015, V-offset: 0.400000, H-pos[seg=0]: -4.94174e-11
chan: 1, nbrSegments: 1, nbrSamplesInSeg: 44000, indexFirstPoint: 2, V-slope: 0.000031, V-offset: 0.000000, H-pos[seg=0]: -4.94174e-11
wf2.shape: (2, 44000) wt2.shape: (2, 44000) |
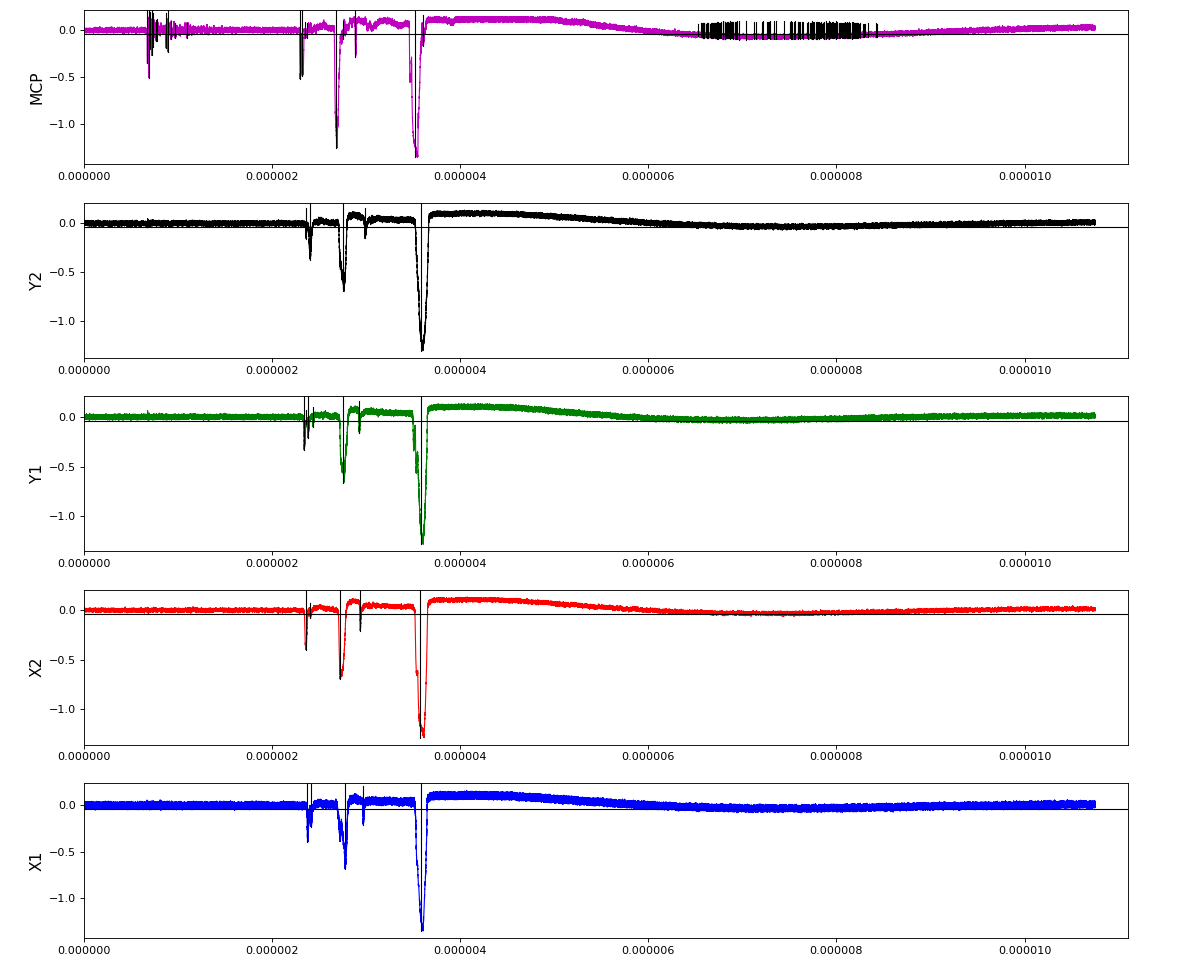
Association of channels of AmoEndstation.0:Acqiris.1
ch = (2,3,4,5,6) - all other channels of Acqiris.1 and 2 are empty or noisy
ylab = ('X1', 'X2', 'Y1', 'Y2', 'MCP')
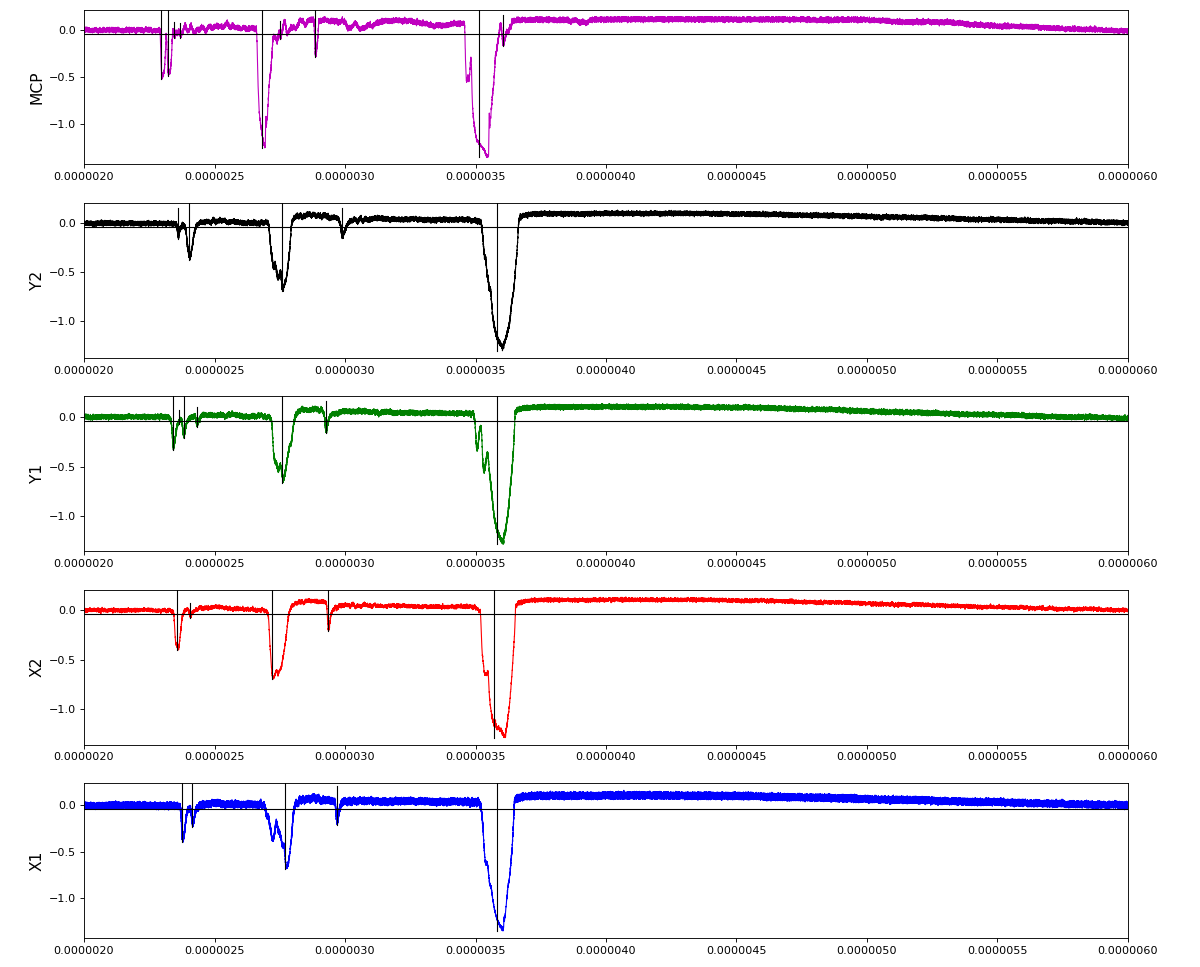
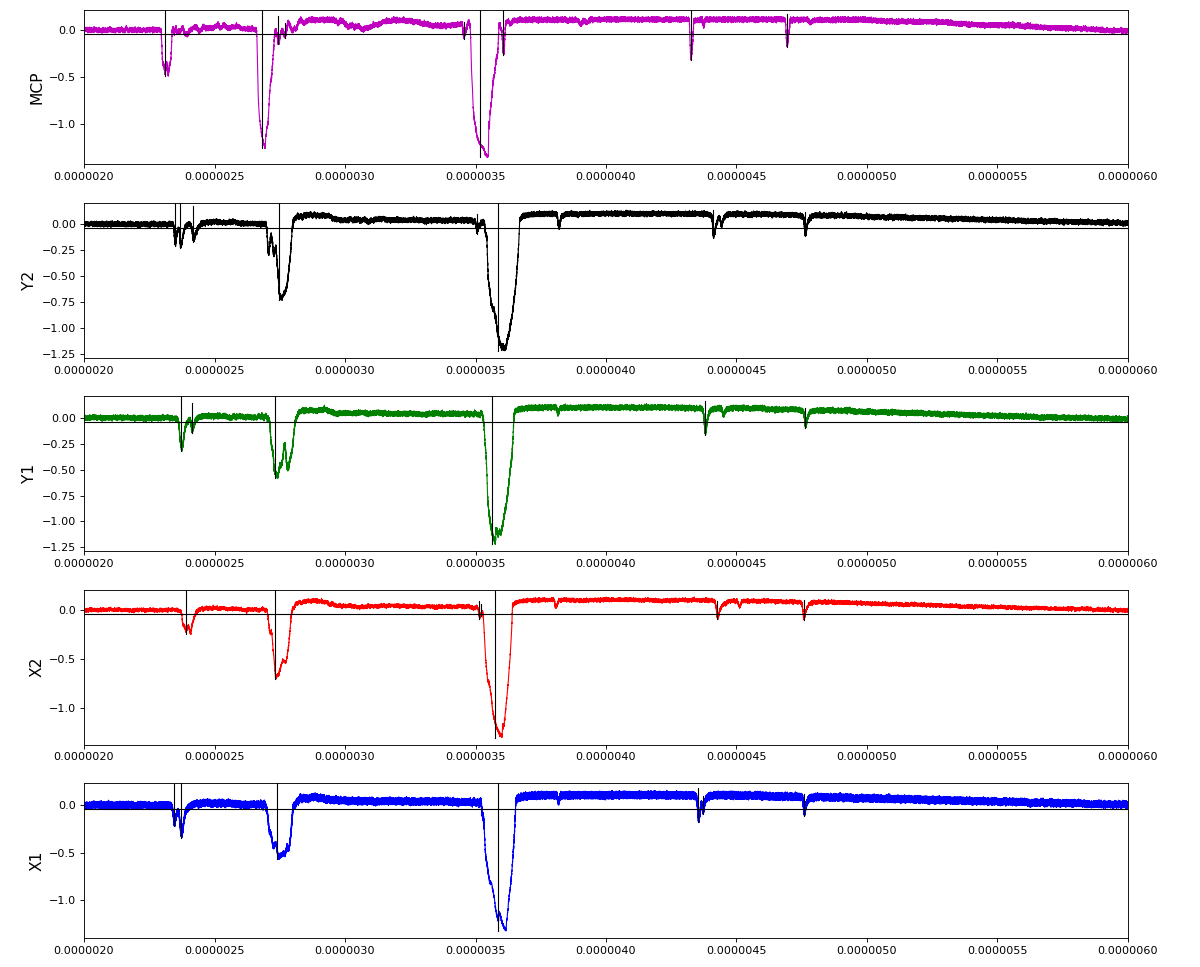
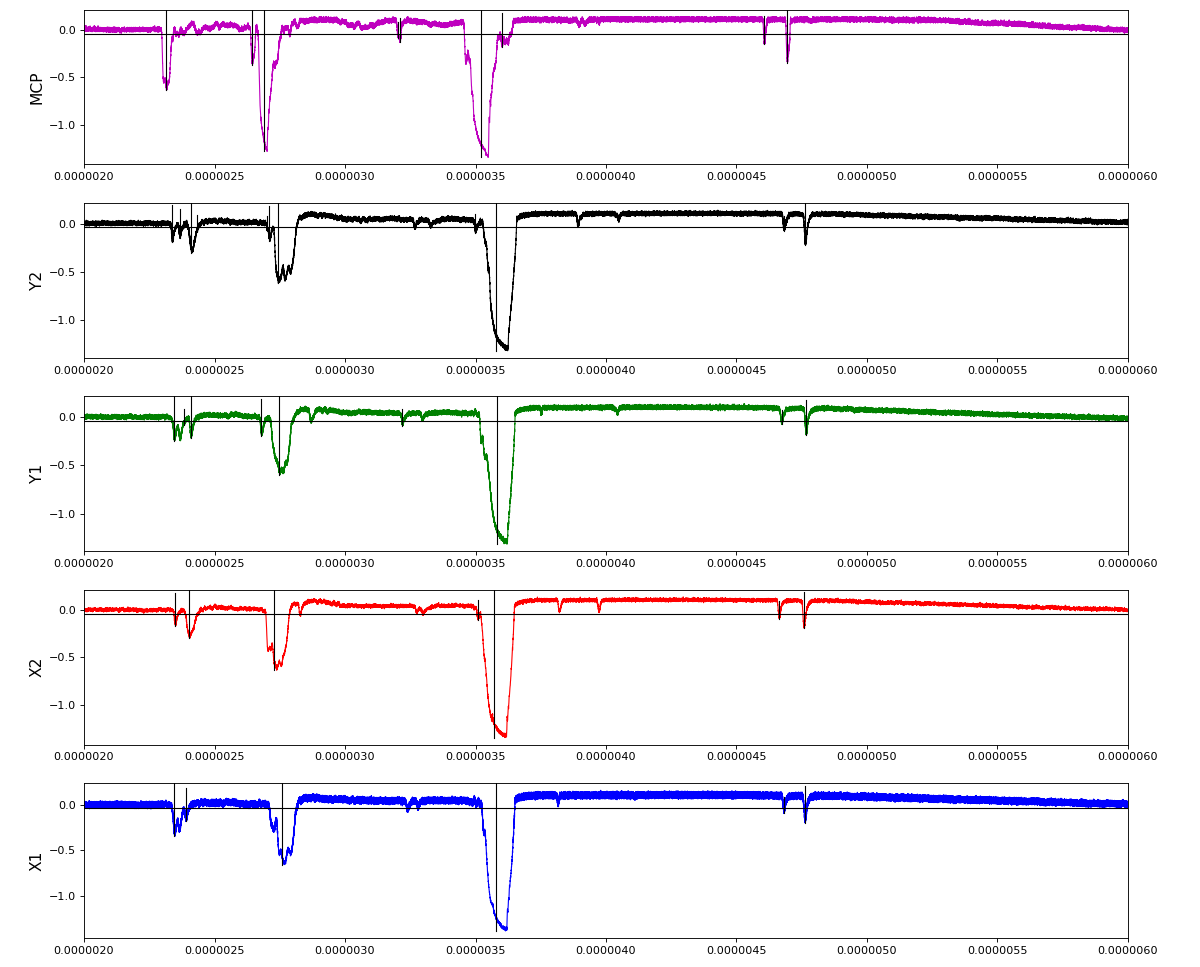
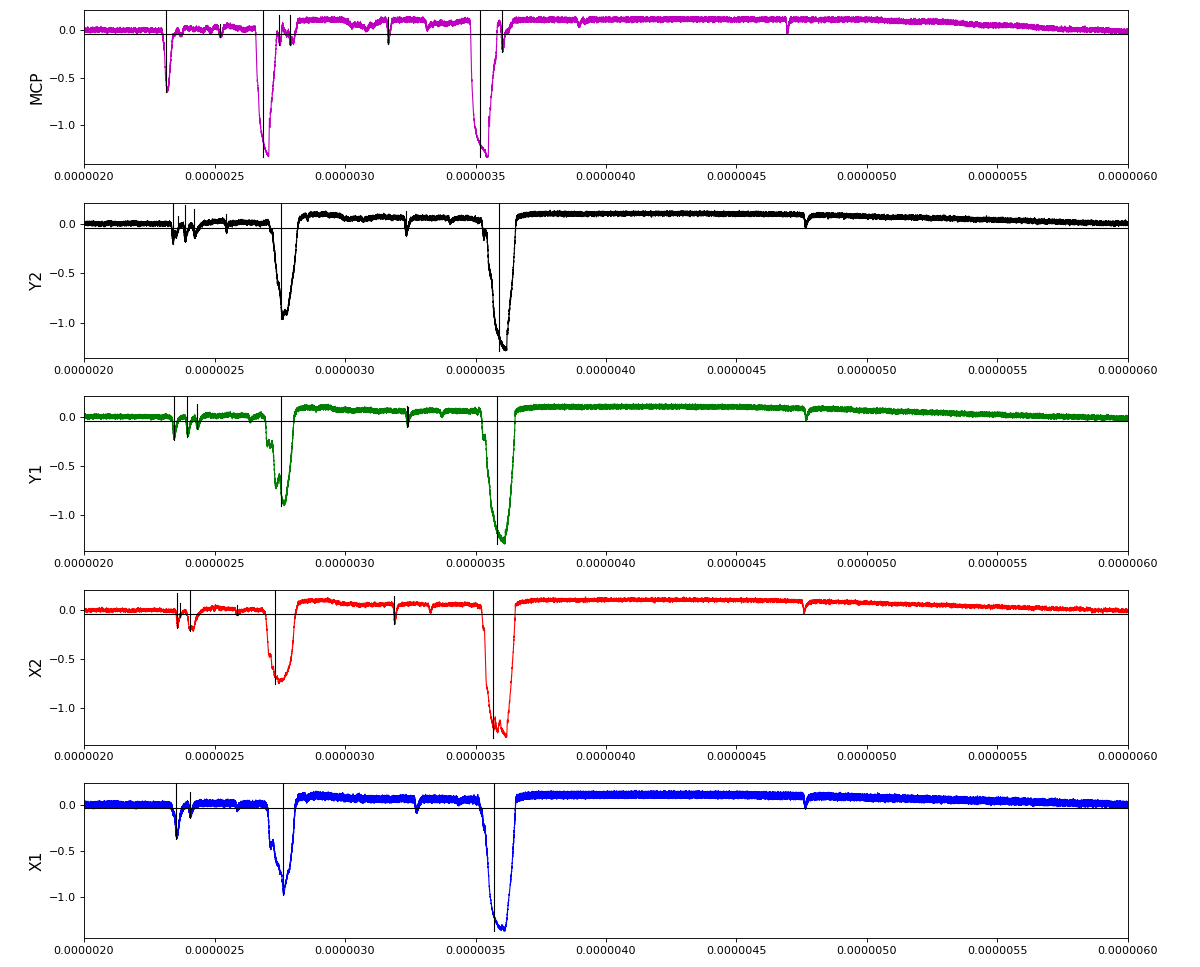
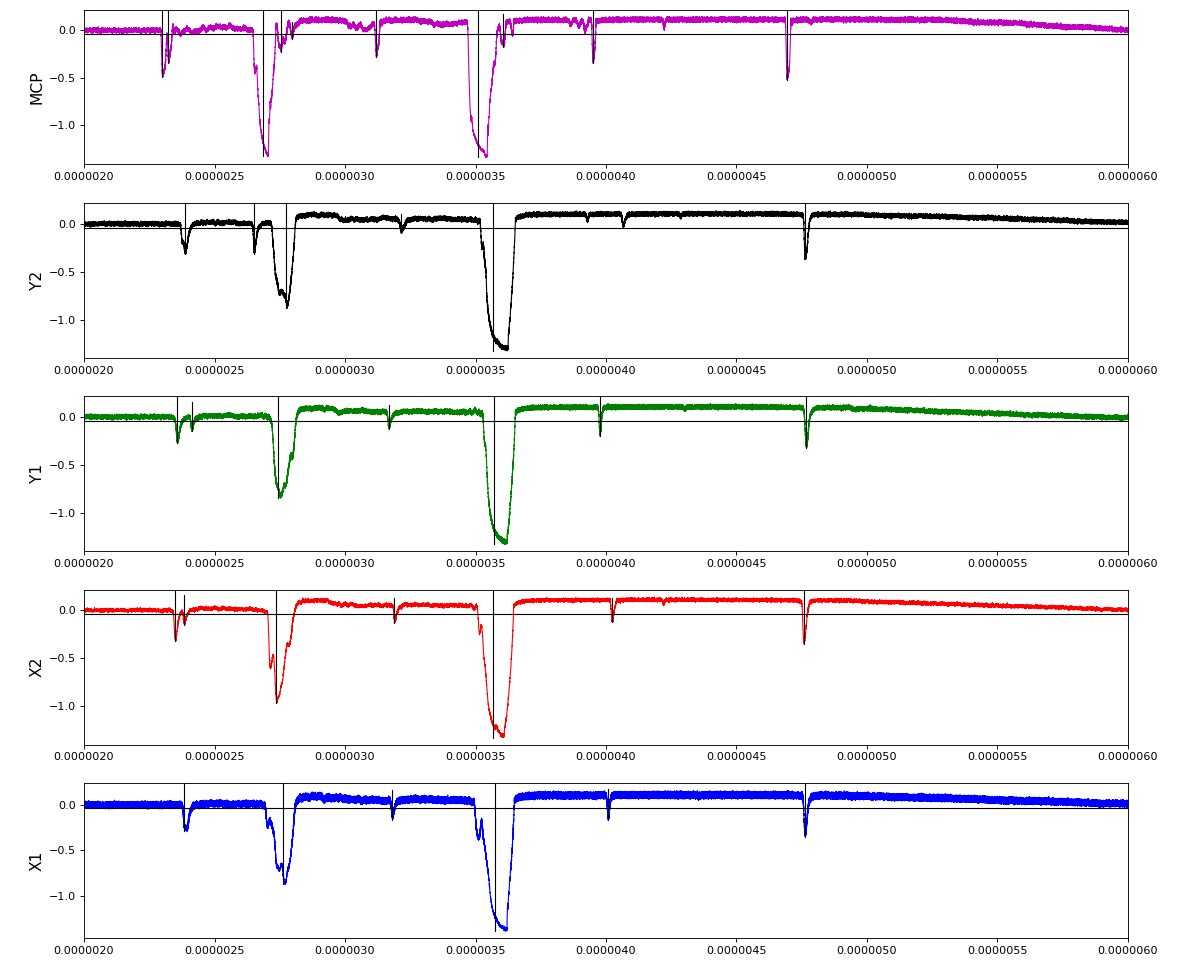
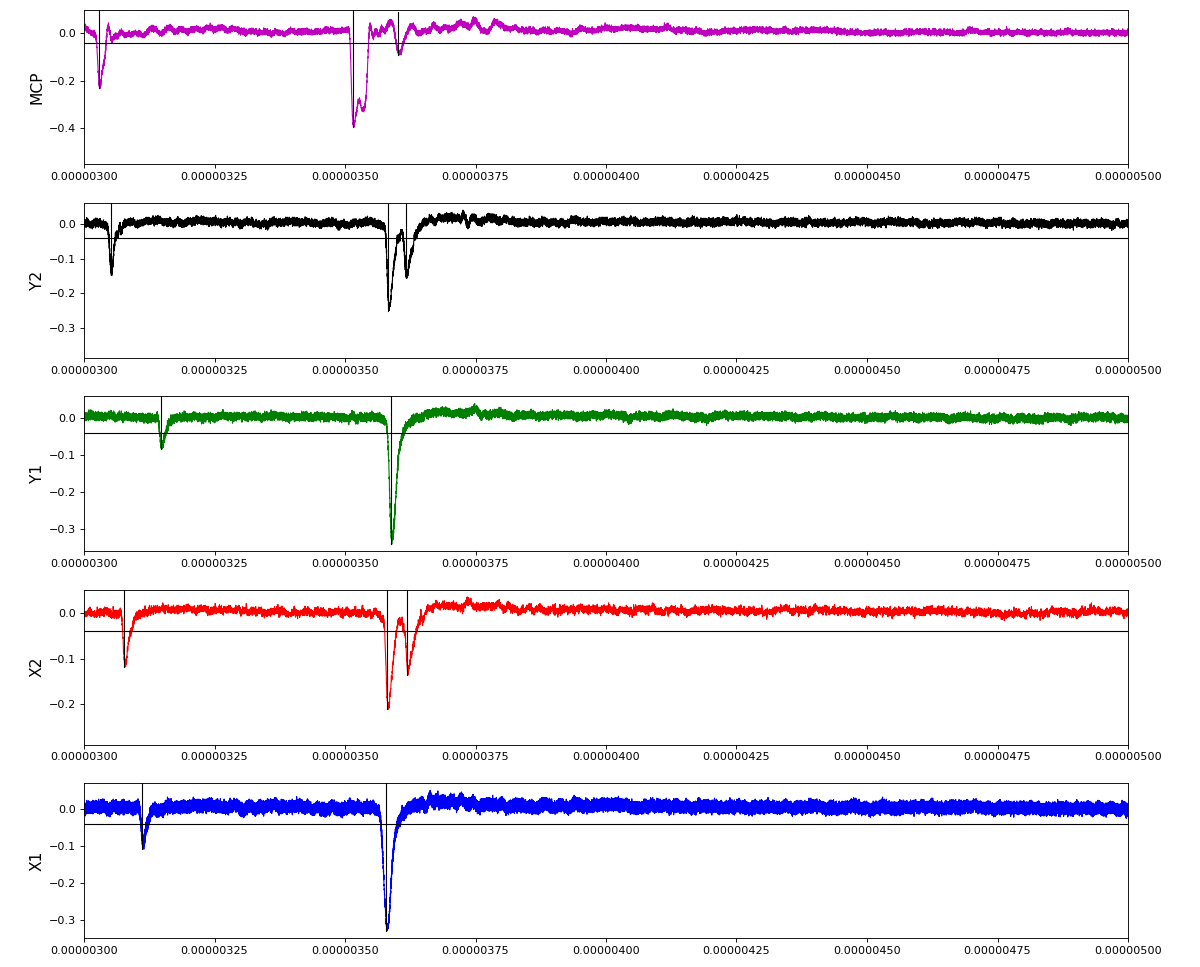
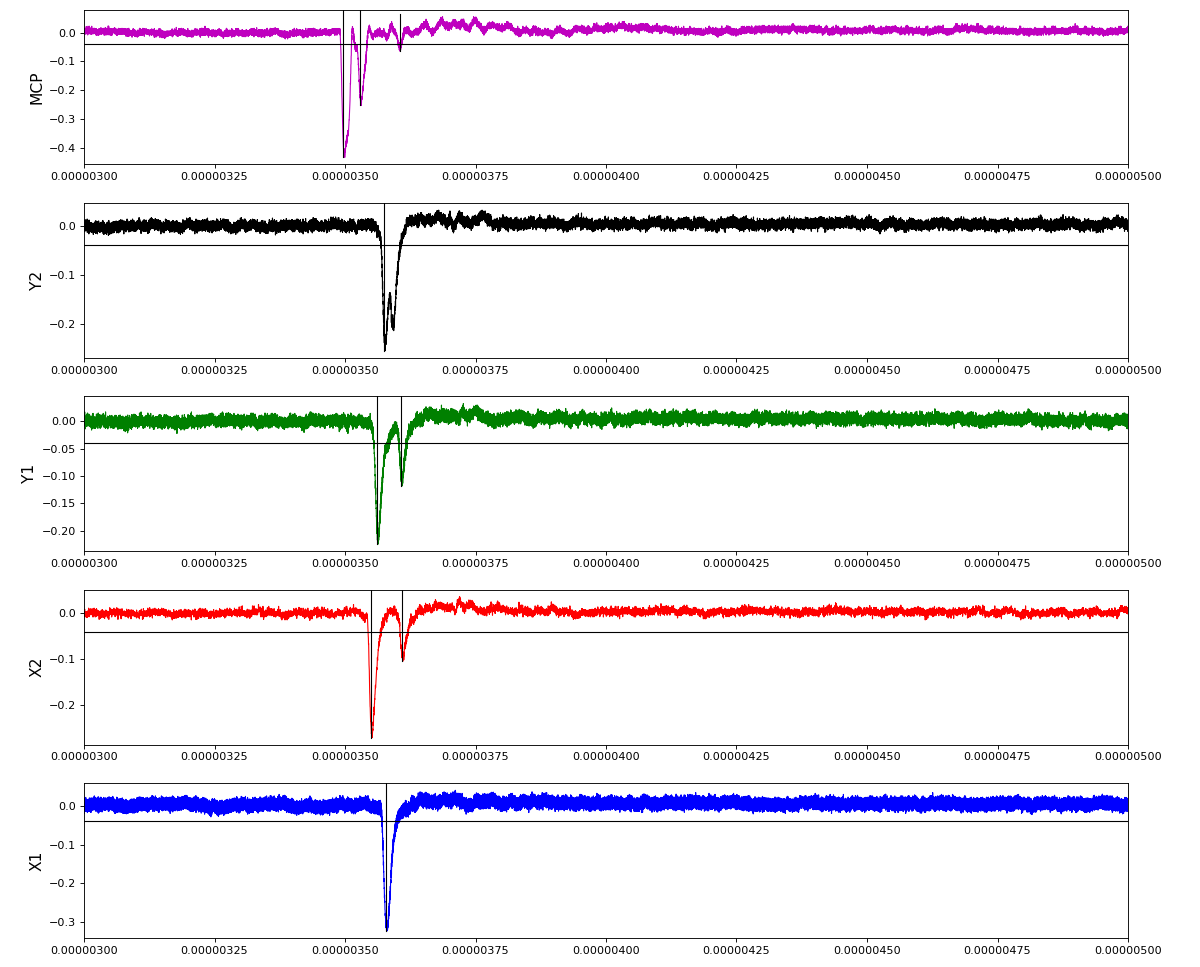
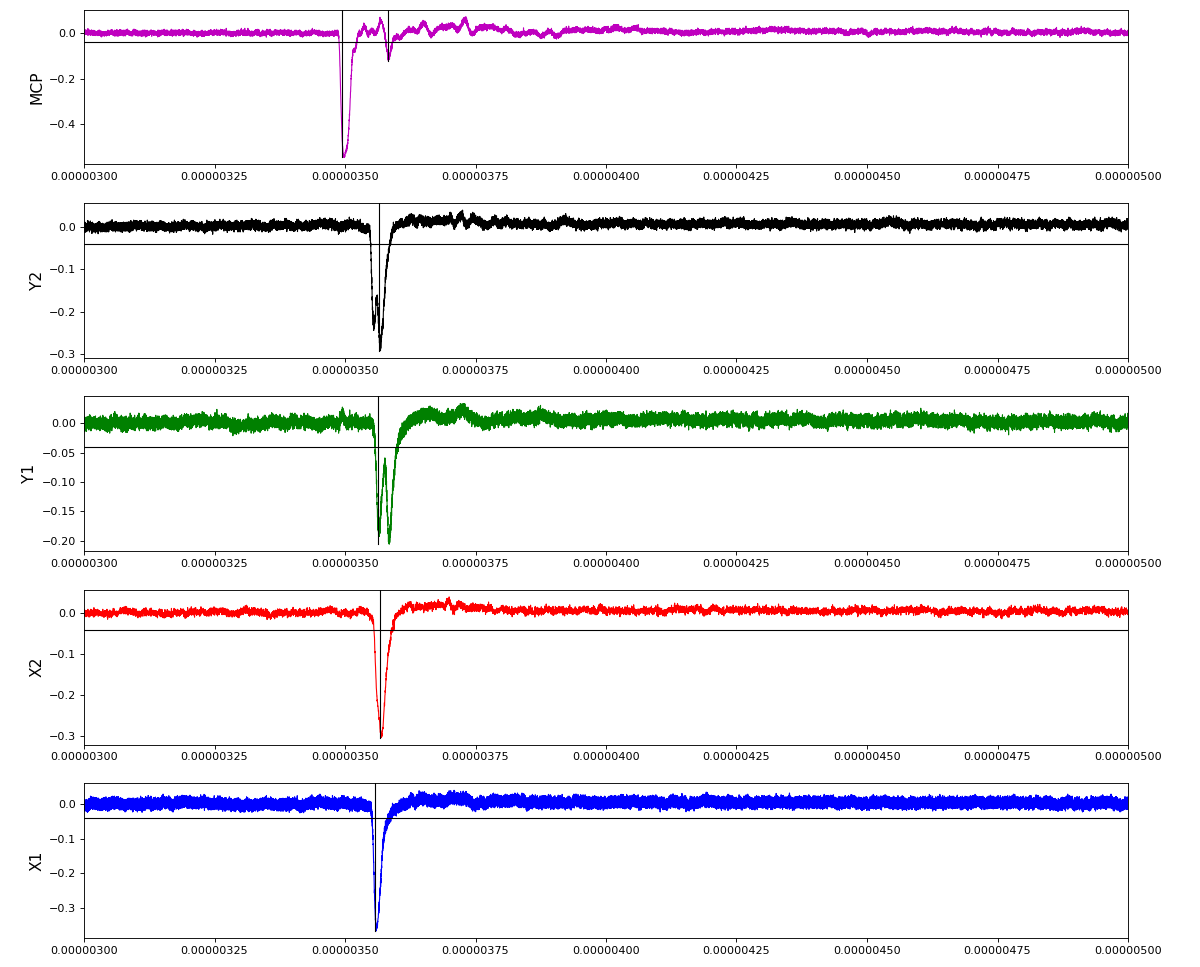
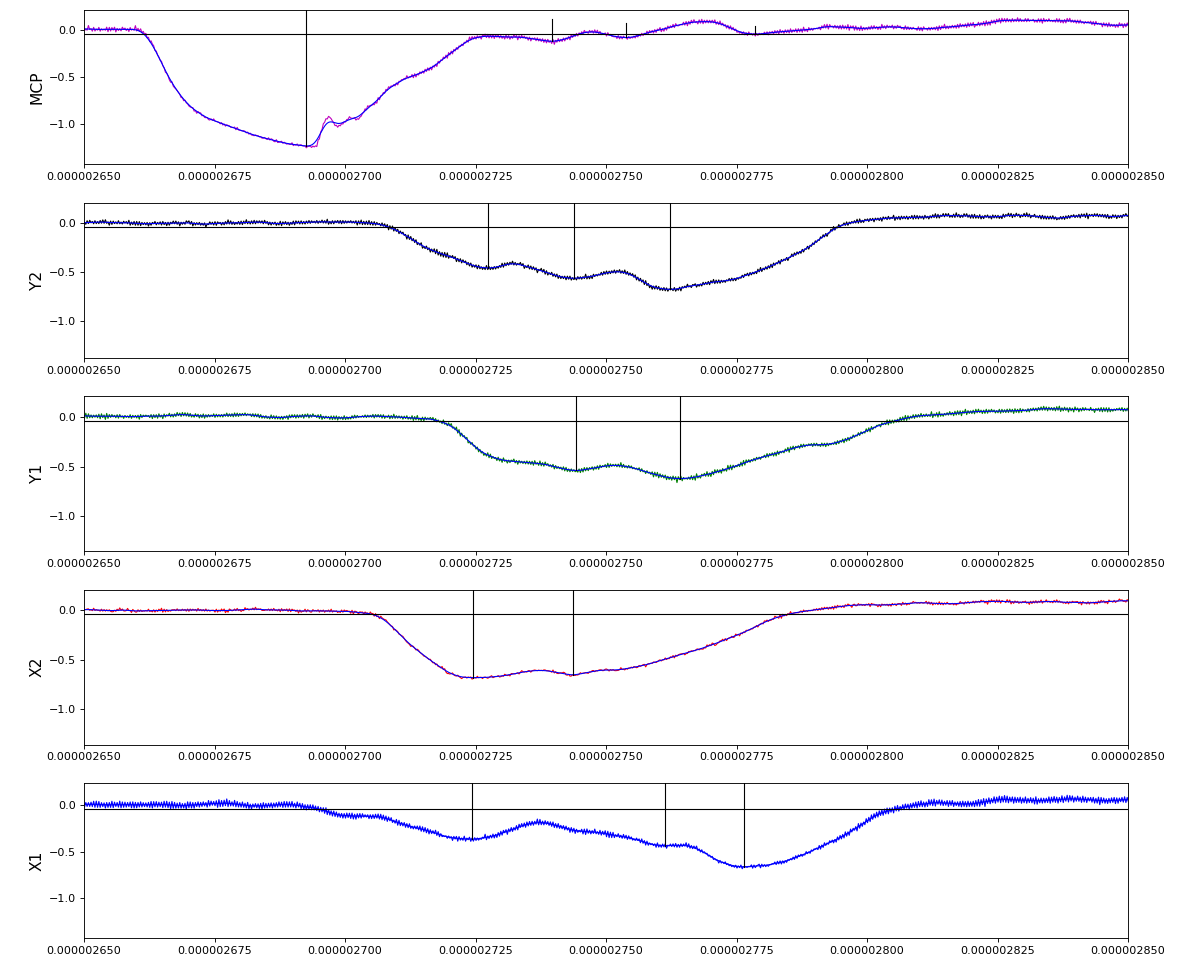
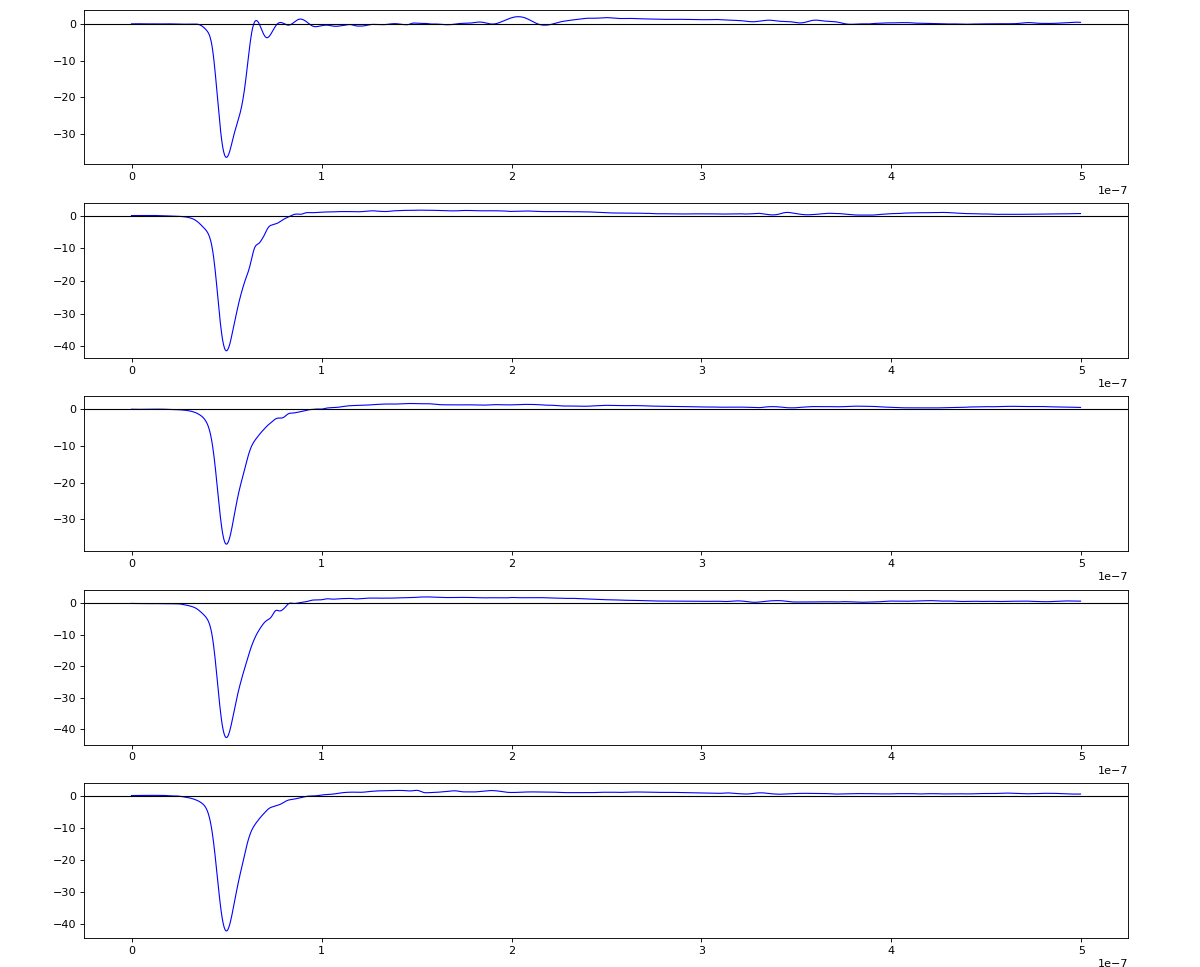
Total waveform with problematic peak reconstruction in MCP channel:
Version of peak finder searching for local extremes after filtering
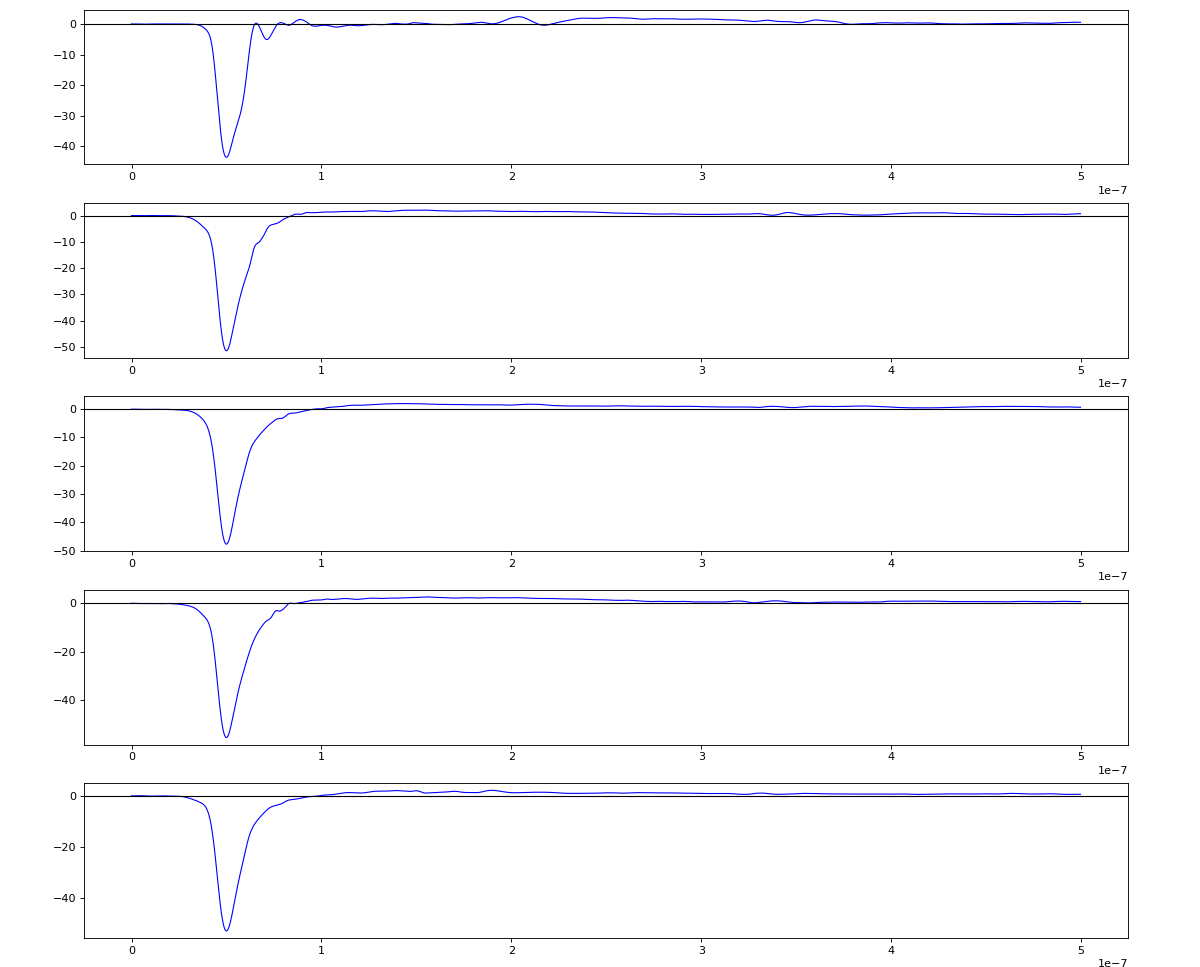
Waveform per channel averaged over 1k raw events with selection of a single peak for amox27716 runs 91,92, and 93
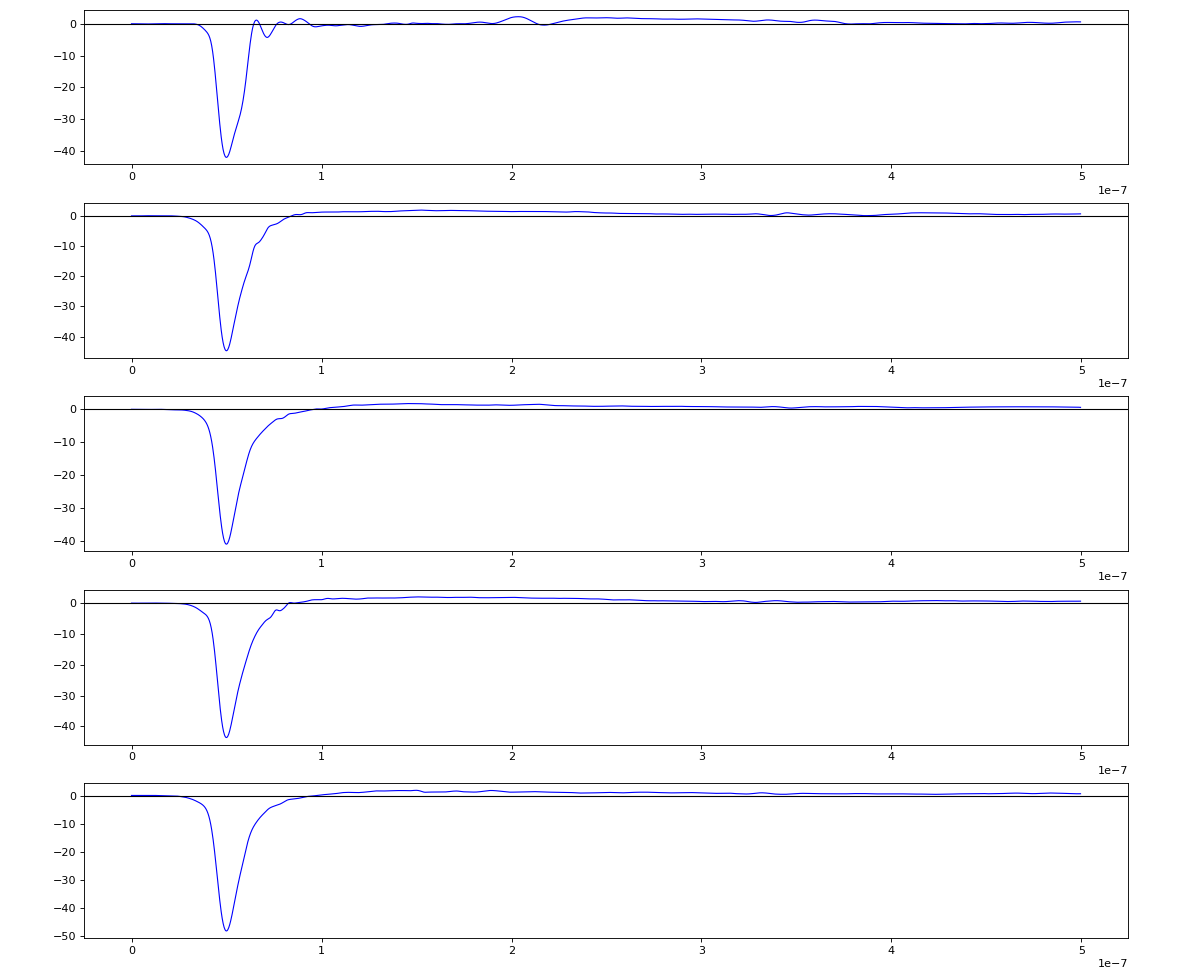
Updated on 2020-10-08
Current hex-/quad-anode calibration procedure is not automated, and needs in expert manual operations...
lcls2/psana/psana/hexanode/examples/ex-24-quad-proc-sort-graph.py two times directly on xtc2 data file for waveform processing and calibration.ex-22-data-acqiris-peaks-save-h5.py or ex-22-data-acqiris-peaks-save-h5-xiangli.py once on xtc2 data file for waveform processing and save intermediate results for number of hits per channel and times per channel in compact hdf5 file. ex-23-quad-proc-sort-graph-from-h5.py two times on hdf5 data file for calibration./reg/g/psdm/detector/data_test/hdf5/amox27716-r0100-e060000-single-node.h5, but real xtc2 data files are not available yet. hexanode/examples/configuration_quad.txtmake sure that the beginning 10 lines of this script are consistent with what you are doing, e.g.
2 // -1 = detector does not exist, 0 = just read (no sort/calib), 1 = sort, 2 = calibrate fu,fv,fw, w_offset, 3 = generate correction tables and write them to disk 0 // hexanode used (yes = 1, no = 0) (Parameter 1101) 0 // 0 = common start, 1 = common stop (for TDC8HP and fADC always use 0) 1 // TDC channel for u1 (counting starts at 1) (Parameter 1129) 2 // TDC channel for u2 (counting starts at 1) (Parameter 1130) 3 // TDC channel for v1 (counting starts at 1) (Parameter 1131) 4 // TDC channel for v2 (counting starts at 1) (Parameter 1132) 0 // TDC channel for w1 (counting starts at 1) (Parameter 1133) HEX ONLY 0 // TDC channel for w2 (counting starts at 1) (Parameter 1134) HEX ONLY 5 // TDC channel for mcp(counting starts at 1) (0 if not used) (Parameter 1135) |
ex-23-quad-proc-sort-graph-from-h5.py set path to the file configuration_quad.txt e.g. ' calibcfg' : '/reg/neh/home4/dubrovin/LCLS/con-lcls2/lcls2/psana/psana/hexanode/examples/configuration_quad.txt',configuration_quad.txtmanually set scale-factor for layer U and V (mm/ns) Parameter 1102, 1103
-133.0 // offset to shift timesum layer U to zero (in nanoseconds) (Parameter 1108) -142.6 // offset to shift timesum layer V to zero (in nanoseconds) (Parameter 1109) 0 // offset to shift timesum layer W to zero (in nanoseconds) (Parameter 1110) HEX ONLY 0. // offset to shift the position picture in X (Parameter 1106) 0. // offset to shift the position picture in Y (Parameter 1107) 5.5 // halfwidth (at base) of timesum layer U (in nanoseconds) (Parameter 1115) 5.5 // halfwidth (at base) of timesum layer V (in nanoseconds) (Parameter 1116) 5.5 // halfwidth (at base) of timesum layer W (in nanoseconds) (Parameter 1117) HEX ONLY 1 // scalefactor for layer U (mm/ns) (Parameter 1102) 1 // scalefactor for layer V (mm/ns) (Parameter 1103) 1 // 0. scalefactor for layer W (Parameter 1104) HEX ONLY 1 // 0. offset layer W (in nanoseconds) (Parameter 1105) HEX ONLY 80. // runtime layer u (in nanosecods) (Parameter 1118) 80. // runtime layer v (in nanosecods) 0 means: same as u (Parameter 1119) 0. // runtime layer w (in nanosecods) 0 means: same as u (Parameter 1120) 40. // radius of active MCP area (in millimeters) (Parameter 1111)(always a bit larger than the real radius) 20. // deadtime of signals from the anode (Parameter 1121) 20. // deadtime of signals from the mcp (Parameter 1122) 1 // use position dependend correction of time sums? 0=no 1=yes (Parameter 1124) 0 // use position dependend NL-correction of position? 0=no 1=yes (Parameter 1125) HEX ONLY |
Achim Czasch <czasch@roentdek.com> Thu 8/15/2019 1:56 AM Hi Mikhail, > Could you remind me please, what is a right order > to adjust parameters and calibrate new detector? yes, there is a sequence that must be followed in the correct order: a) First you must set the parameters in the config file so that the time sum peaks get shifted to zero. b) Then you set the 'runtime'. c) Then you set the xy calibration factors so the that image gets the size that you think is right. (and set the xy offset parameters so that the image is well centered.) d) Then you set 'radius'. This value should be 1 or 2 mm larger so that it really includes all hits. best, Achim ================================== Achim Czasch <czasch@roentdek.com> Fri 8/16/2019 12:00 AM Hi Mikhail, > Scale factors converting time to coordinate are not important now. > because I am not aware about precise detector geometry. For the DLD (not HEX) you must set the 2 calibration factors manually later. Example: If you have MCPs with 40mm active diameter then you must the factors so that the image has a diameter of 40mm. The factors for x and y will be slightly different to achieve a round image. > >> b) Then you set the 'runtime'. > Could you remind me please, how can I get this value(s)? You plot x1-x2 (units: ns) in a 1D-plot. Then logy-scale. Now you can see a distribution that ranges from -z to +z (z is just a number here). For 'runtime' you chose z + 2 ns. > But, distributions of the time_sum for u and v look flat, even without calibration tables. The errors that are corrected are within about +-4 ns. So maybe you must zoom in a bit (in the y-scale). But for a DLD this correction not so important. > Still, it looks important to have an ability to calibrate potential differential non-linearity > of the delay lines. I hope this is possible for QUAD-anode, right? Not as easily as with the HEX. With a HEX we have a 3rd layer. This layer provides the additional info that we need to make an automatic linearity calibration of the 3 delay lines. But with a normal DLD we do not have enough information. So this algorithm can not be applied. You can use a pin hole mask and measure the non-linearity manually. Then you can apply another code from us that will spit out correction tables. But this process is very very tedious and time consuming - and if you change something (cable lengths, even voltages) then you may have to do it again. best, Achim |
edit file configuration_quad.txt and set the 1-st parameter "command" to 3.
edit script like ex-23-quad-proc-sort-graph-from-h5.py and set pass where time correction table will be saved, e.g. 'calibtab' : '/reg/neh/home4/dubrovin/LCLS/con-lcls2/lcls2/psana/psana/hexanode/examples/calibration_table_data_new.txt'
run again script ex-23-quad-proc-sort-graph-from-h5.py
check results in calibration_table_data_new.txt
edit file configuration_quad.txt
set the 1-st parameter "command" to 1.
set Parameter 1124 in configuration_quad.txt
Both, configuration and calibration files need to be saved in calibration DB for automated data processing. This operation needs in actual experiment and detector names, run, timestamp, etc. Preliminary deployment commands:
cdb add -e amox27716 -d tmo_quadanode -c calibcfg -r 100 -f configuration_quad.txt -i txt -u dubrovin cdb add -e amox27716 -d tmo_quadanode -c calibtab -r 100 -f calibration_table_data.txt -i txt -u dubrovin |
Content of the DB can be explored with GUI started by command calibman.
See examples ex-24* or ex-25*.
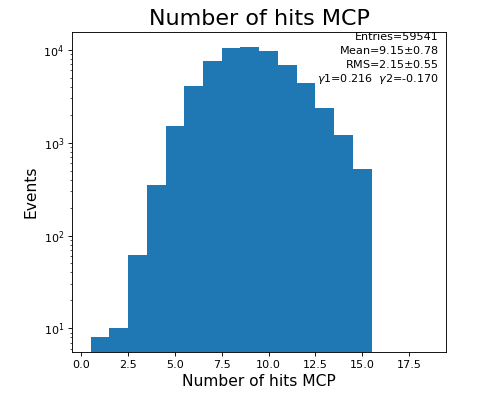
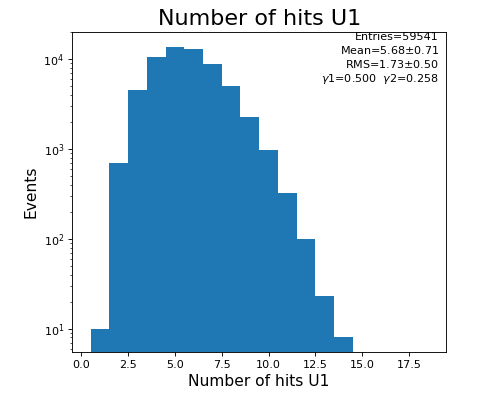
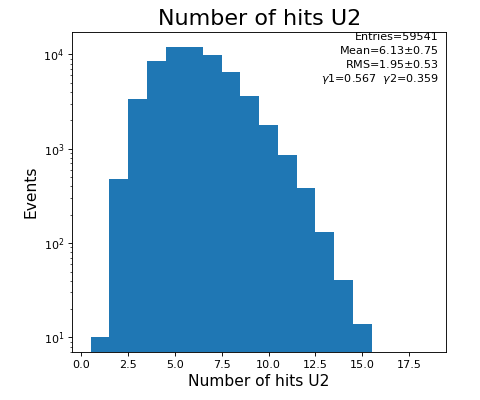
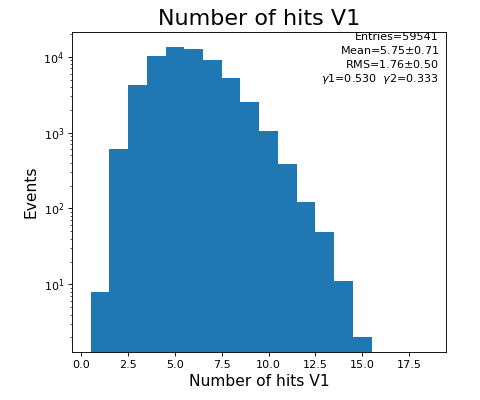
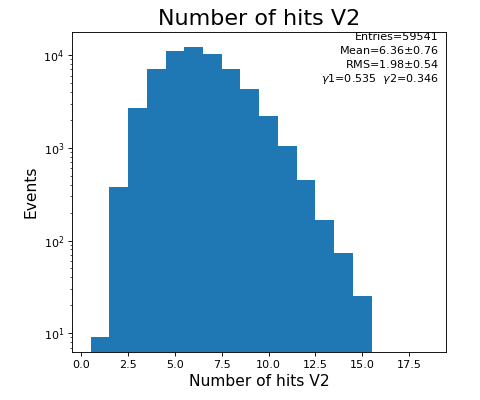
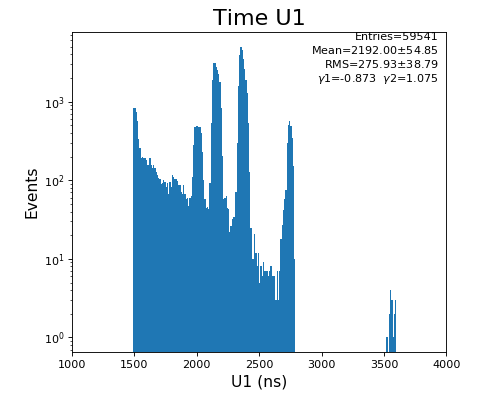
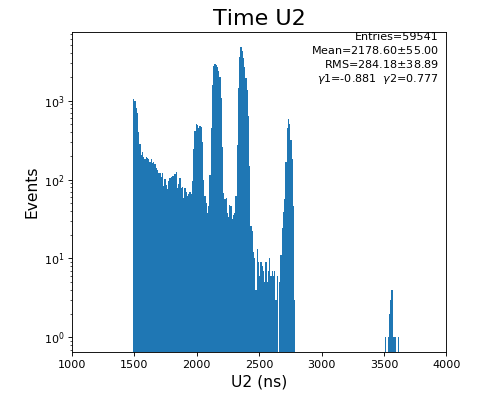
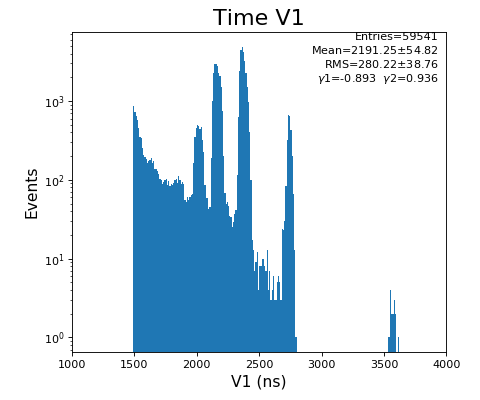
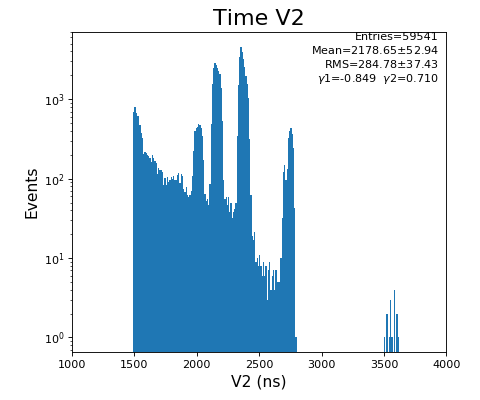
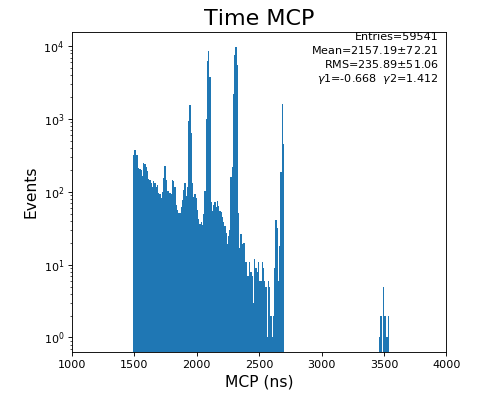
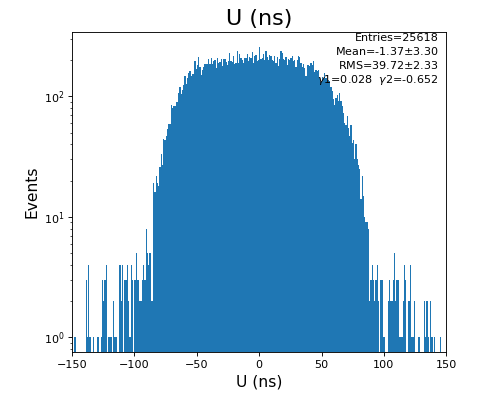
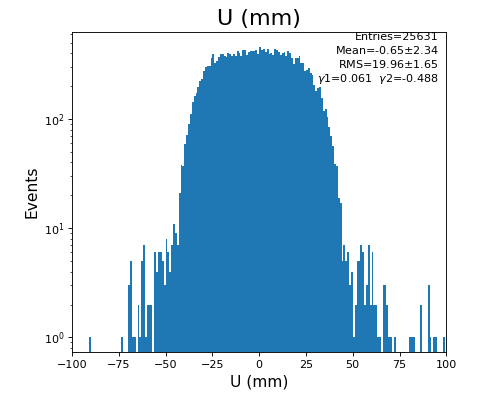
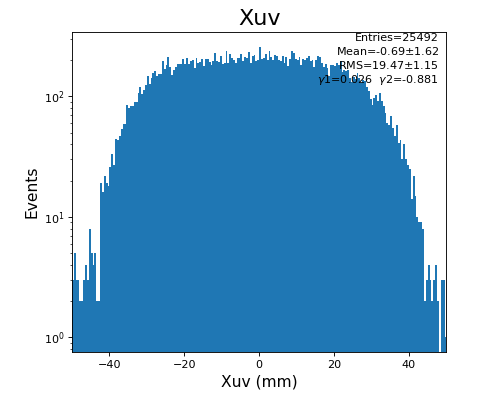
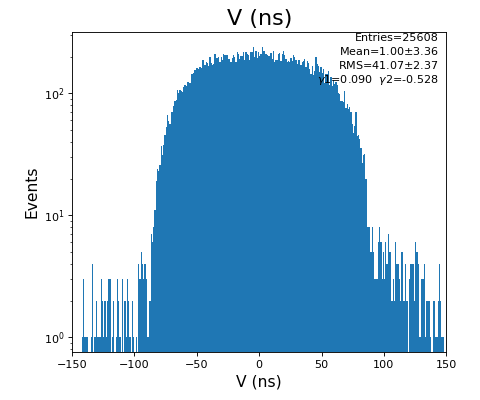
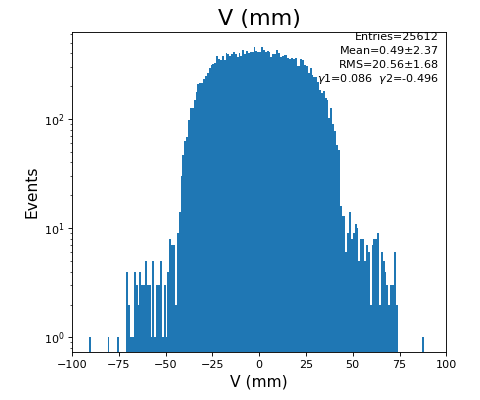
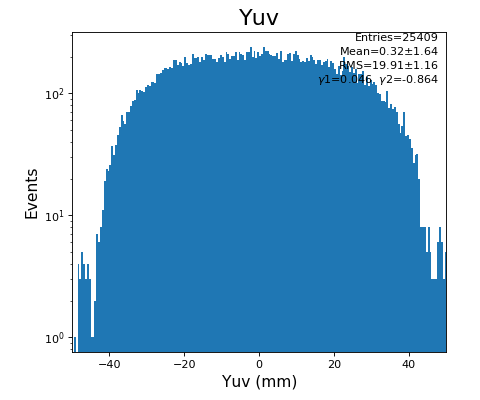
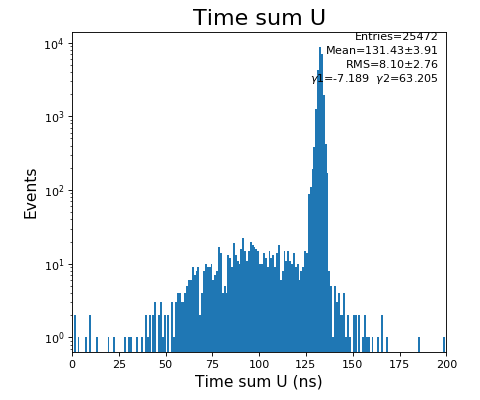
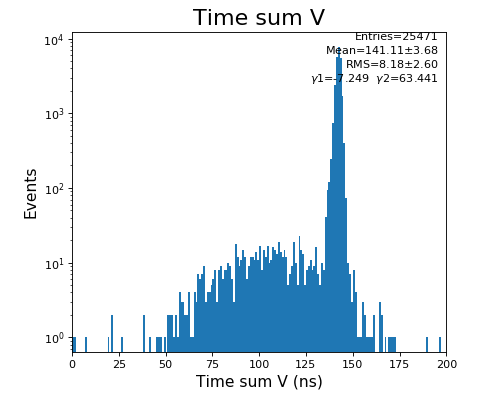
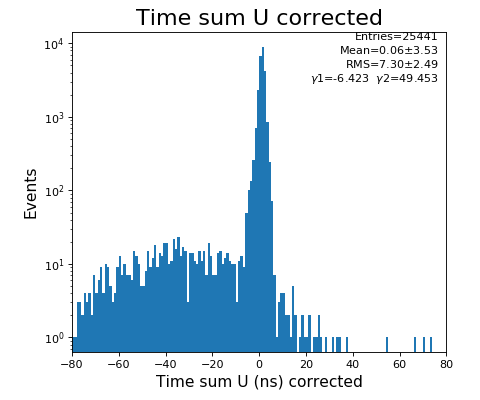
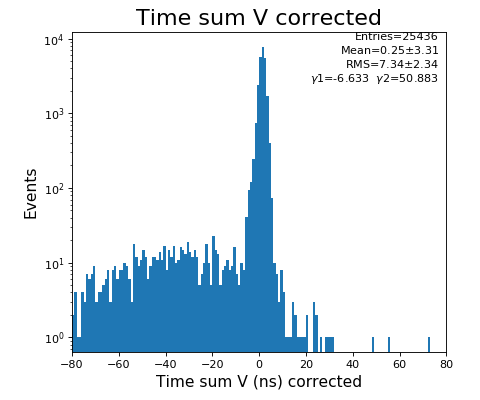
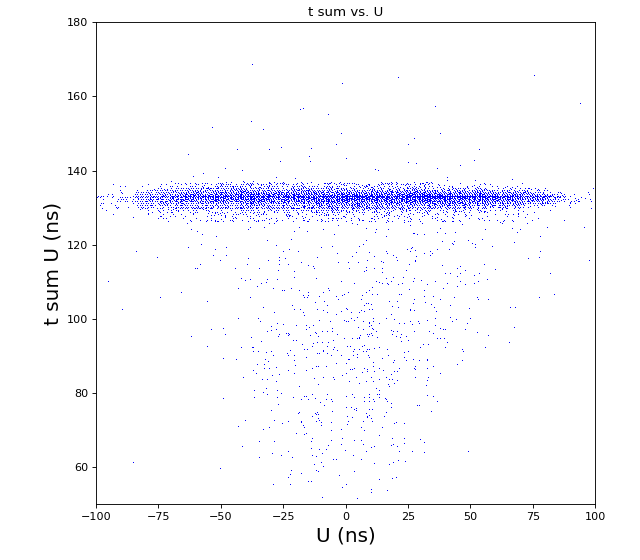
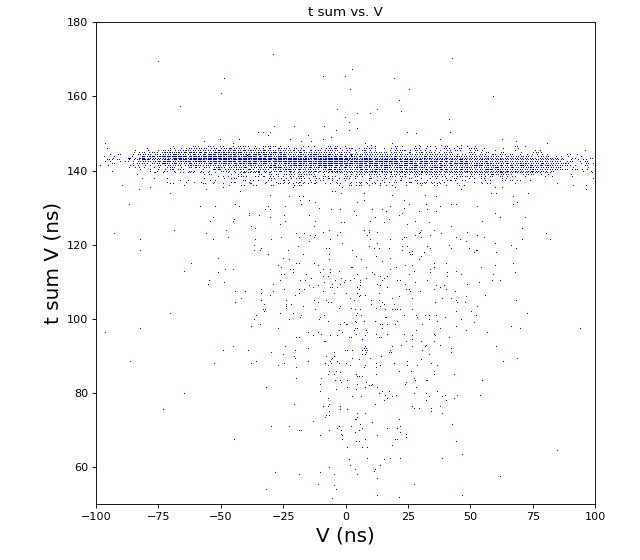
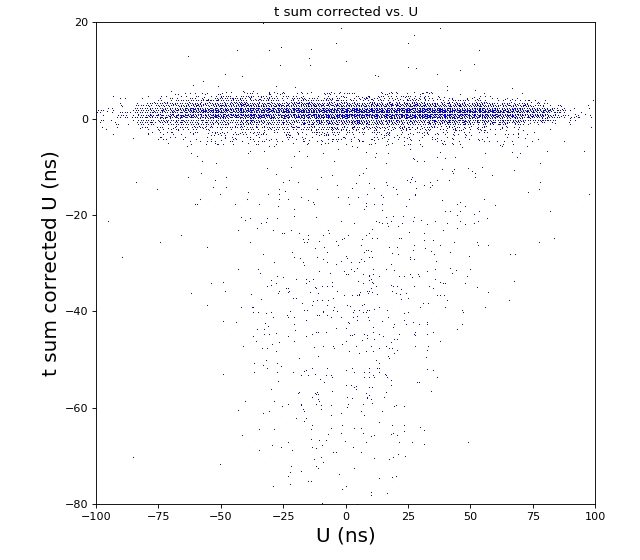
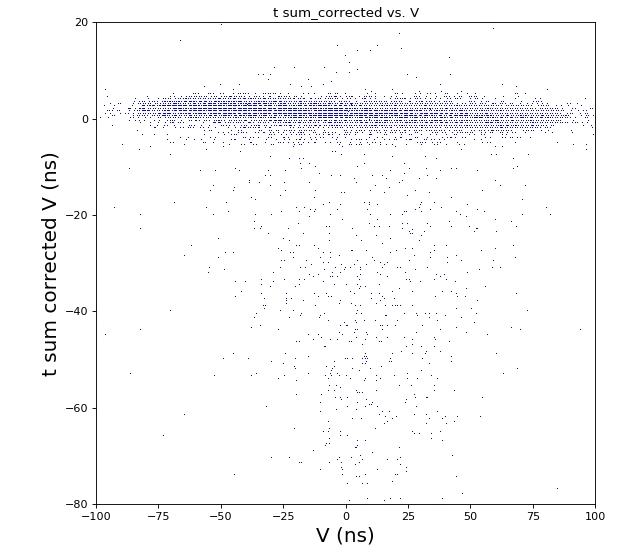
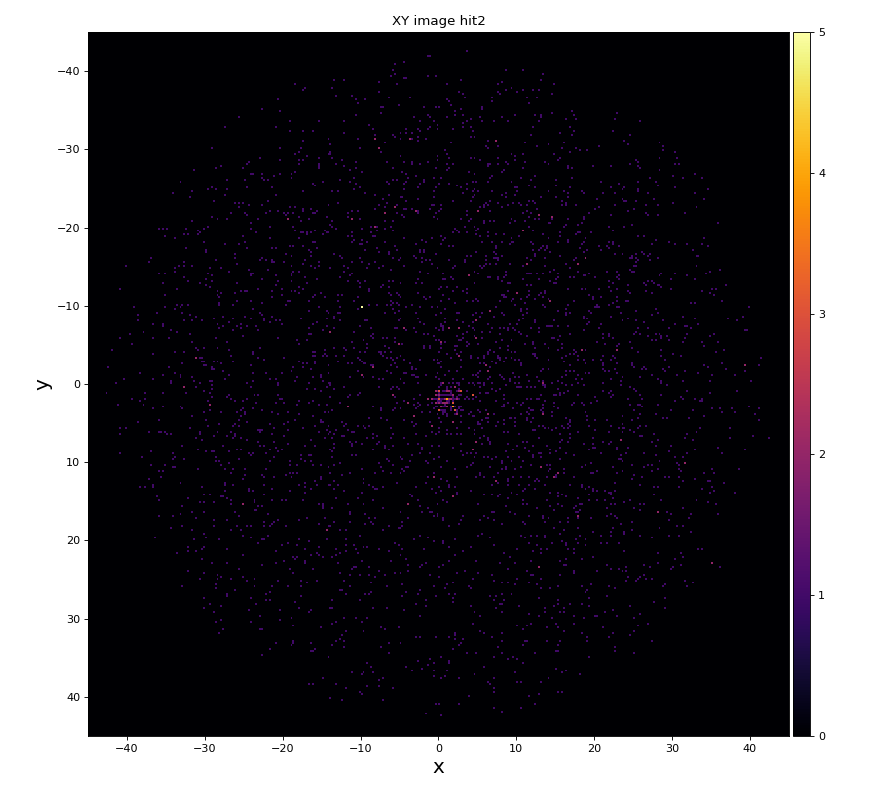
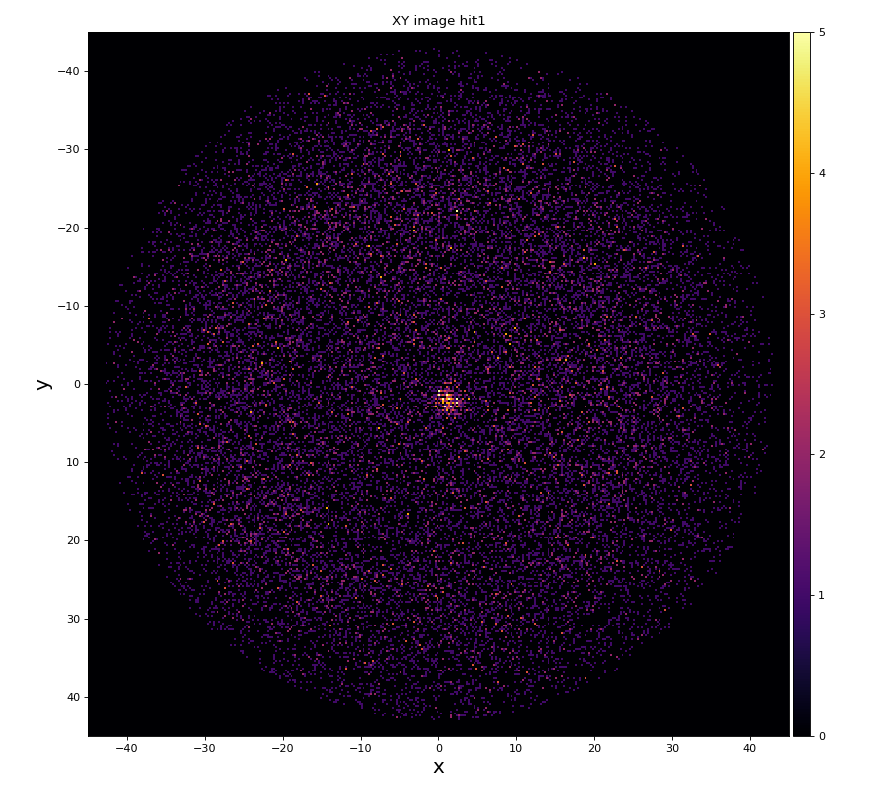
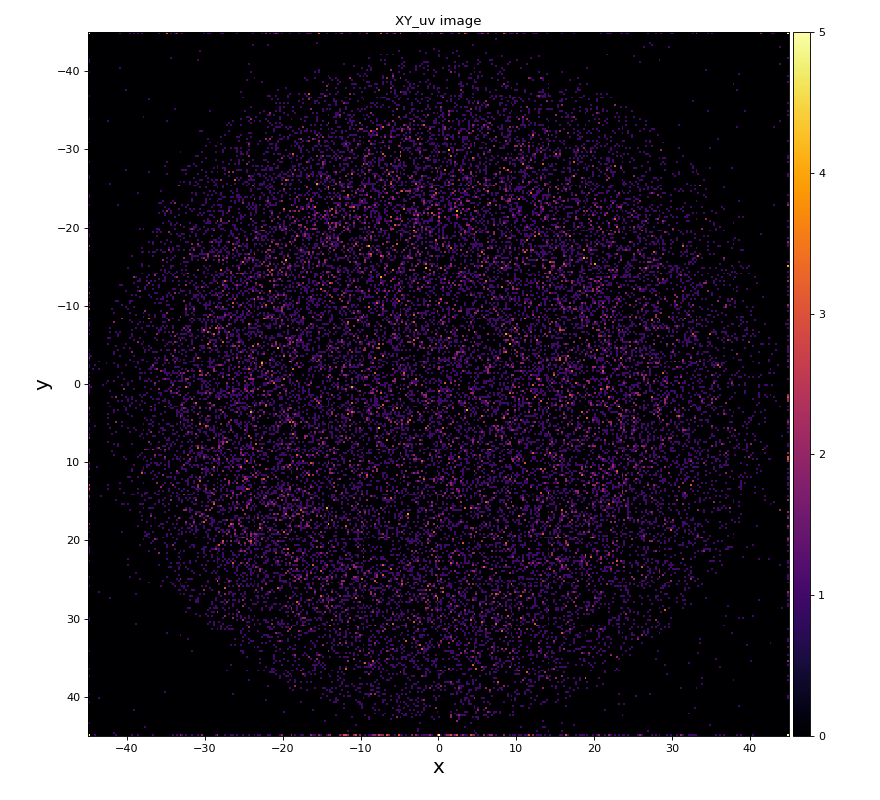
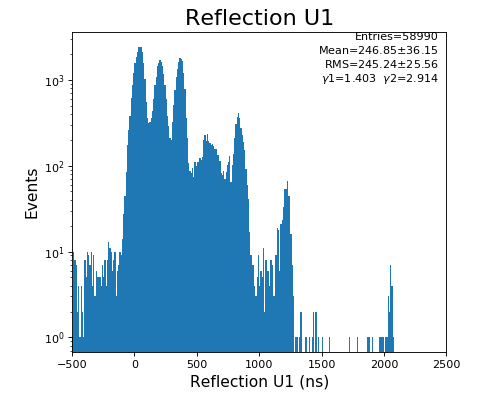
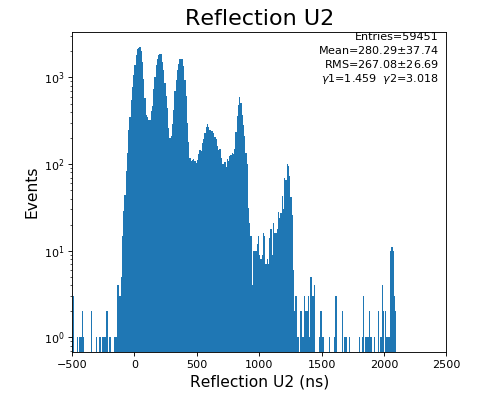
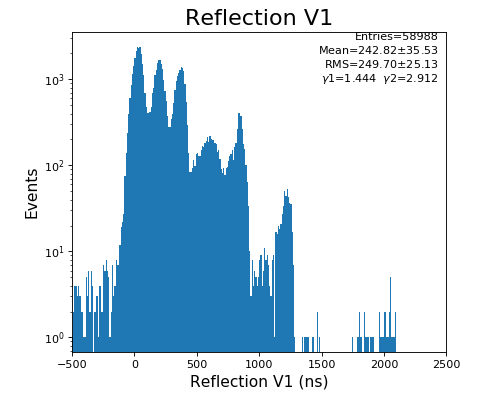
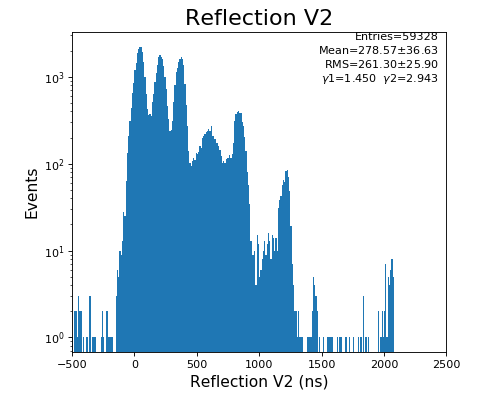
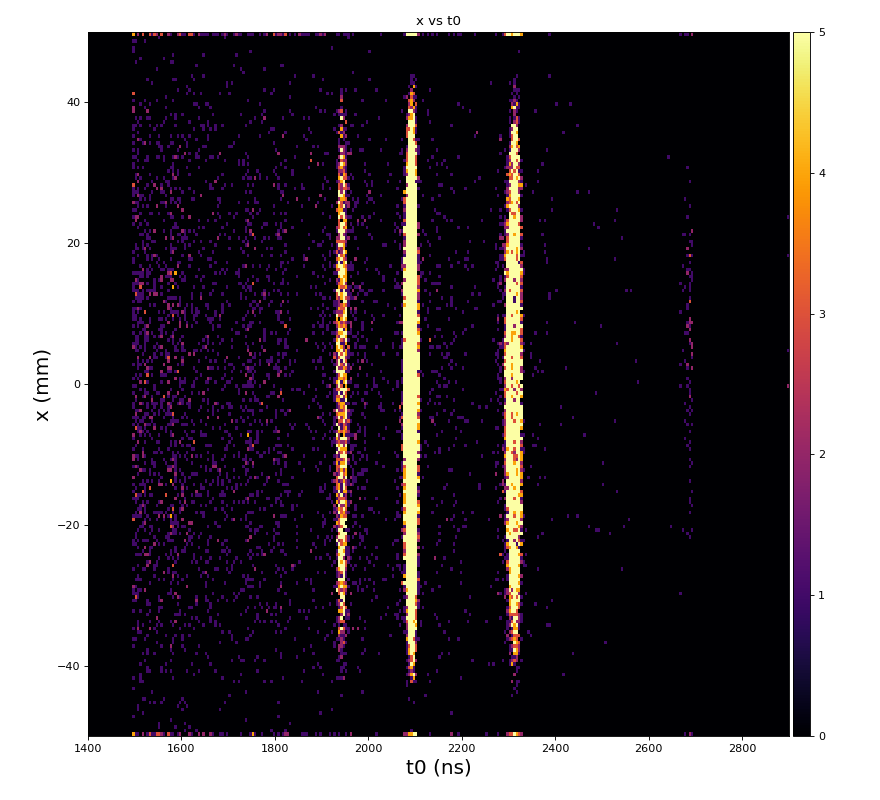
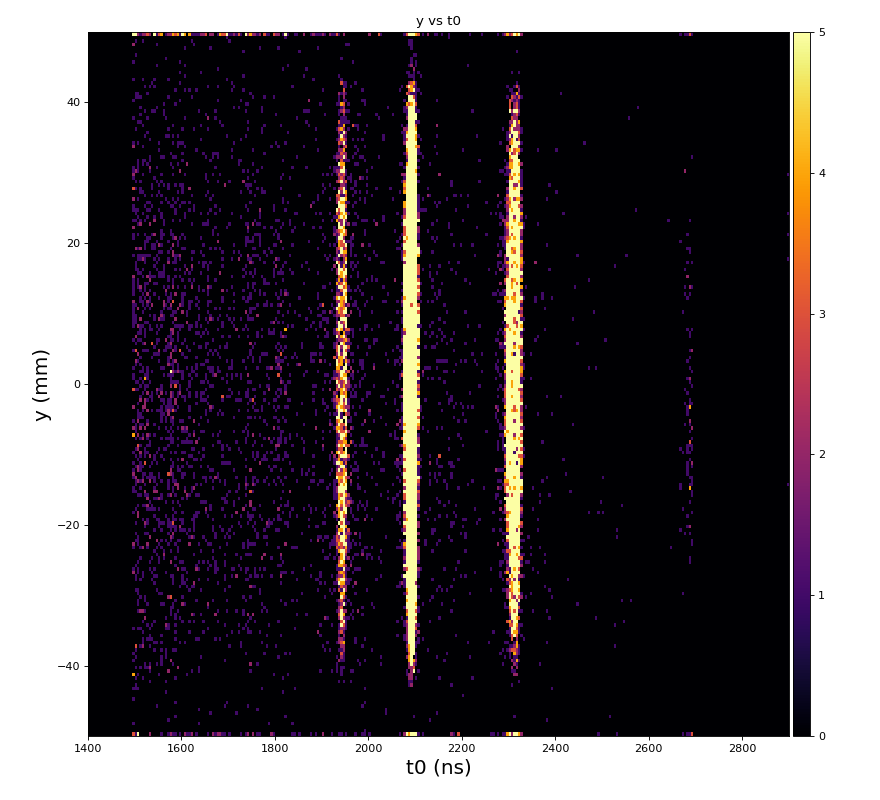
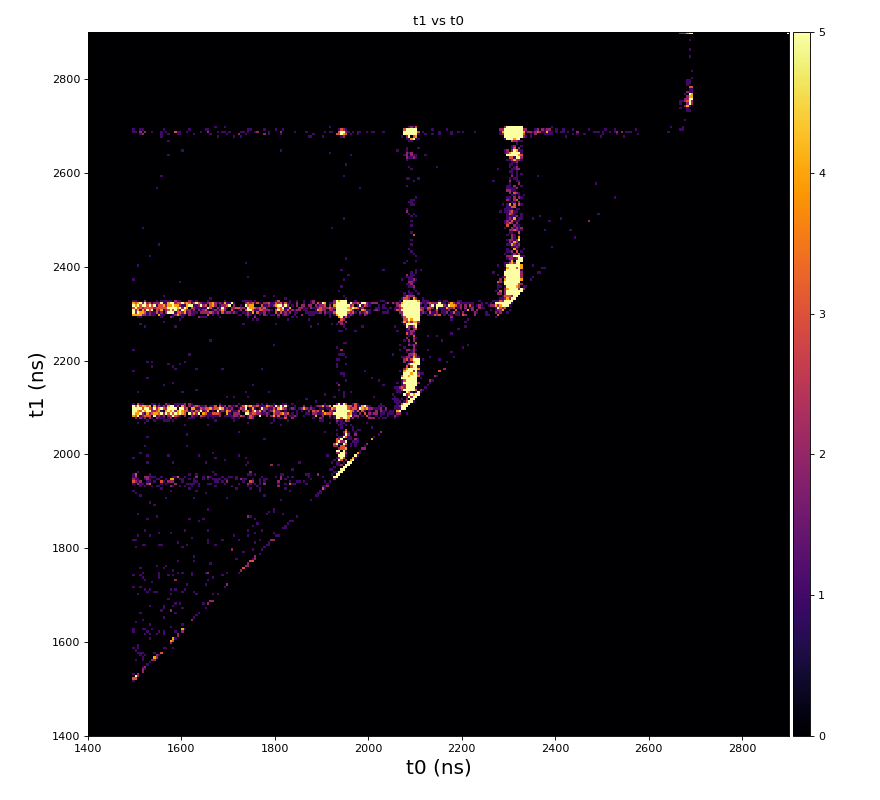
hexanode/examples/ex-quad-09-sort-graph-data.py with command 1 (after calibration command 2,3) set in configuration_quad.txt.
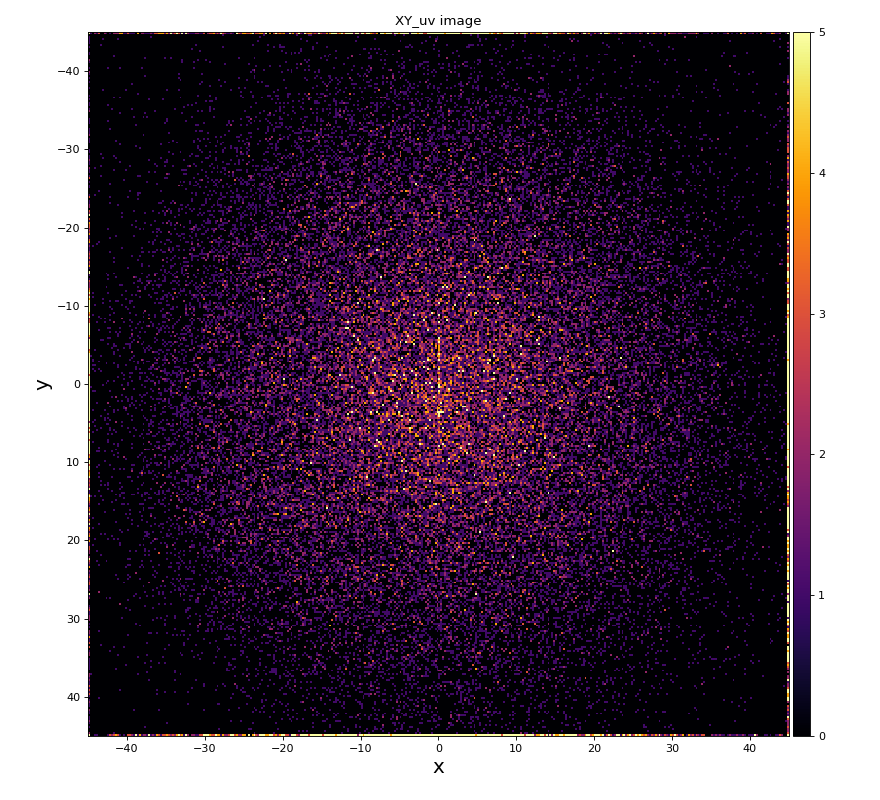
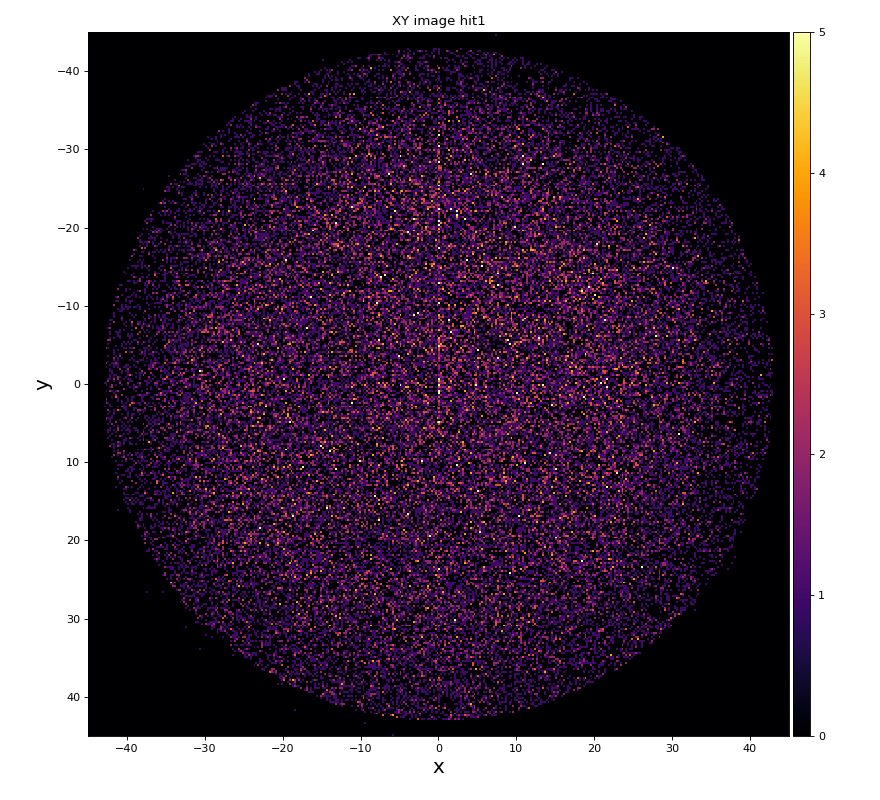
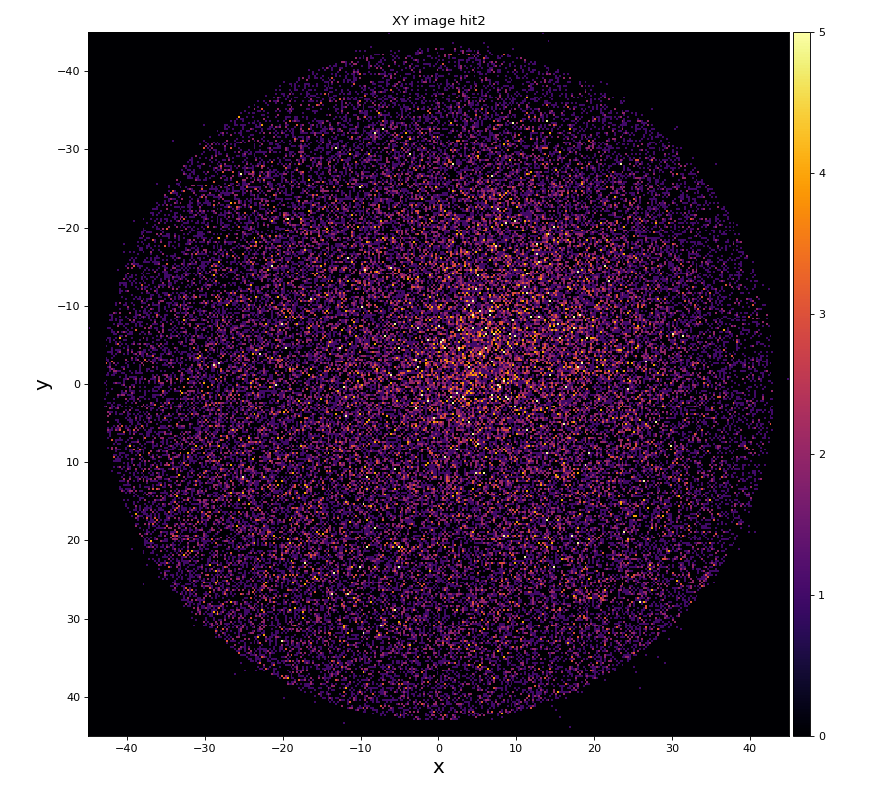
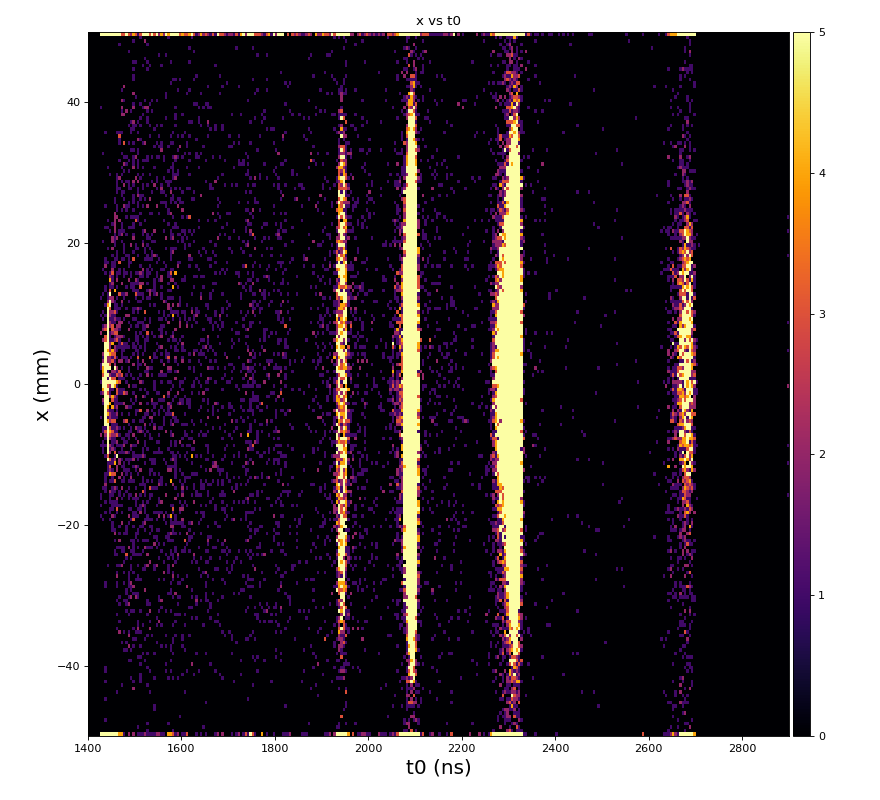
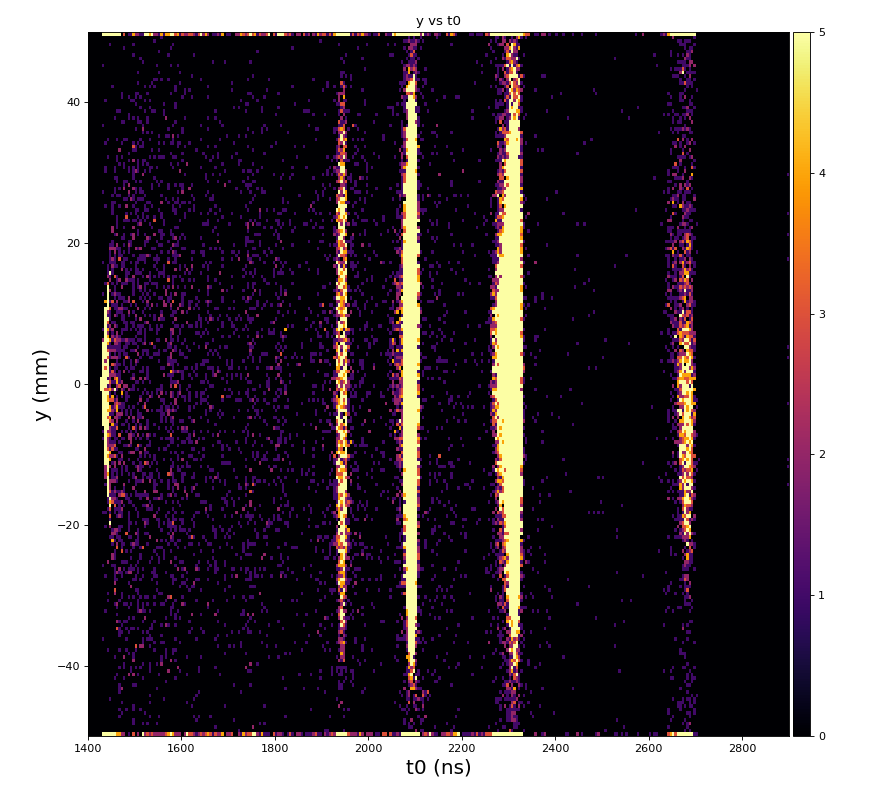
https://github.com/slac-lcls/lcls2/blob/master/psana/psana/hexanode/examples/
ex-20-data-acqiris-access.py - access to detector waveforms
ex-21-data-acqiris-graph.py - plot waveforms and found peaks
ex-22 - 24 - intended for calibration and representative graphics
ex-25-quad-proc-data.py - reads waveforms from xtc2, find peaks, and reconstruct hits using Roentdec library
From ex-23-quad-proc-sort-graph-from-h5.py or ex-24 if xtc2 file is available:
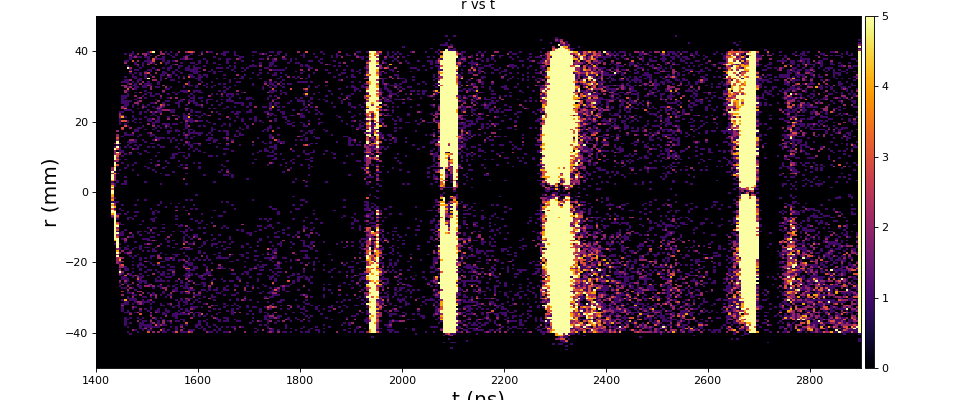
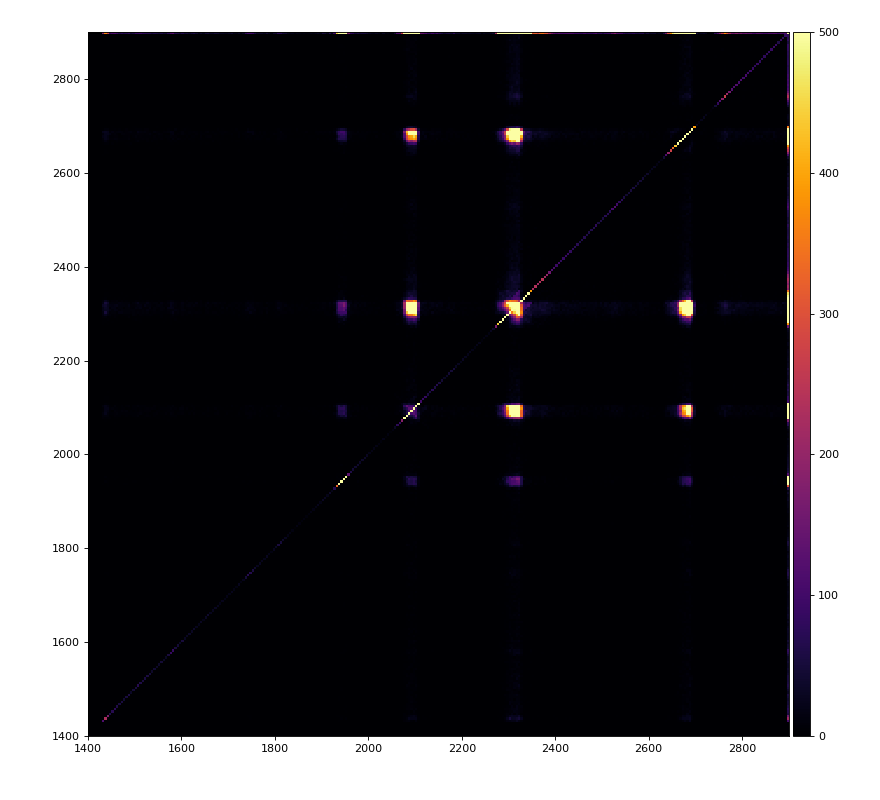
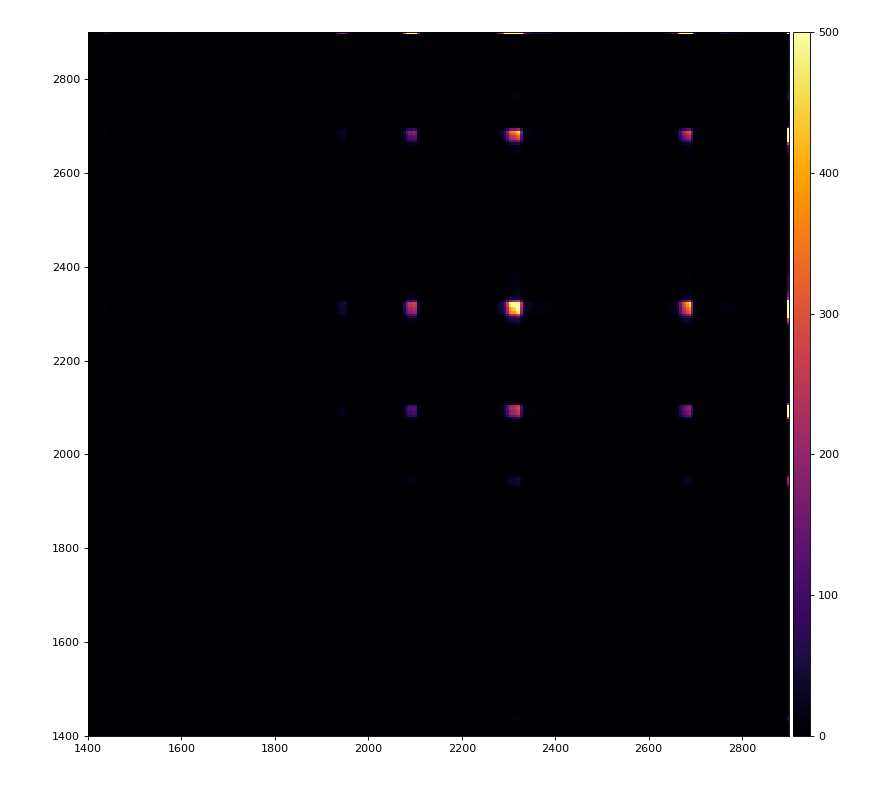
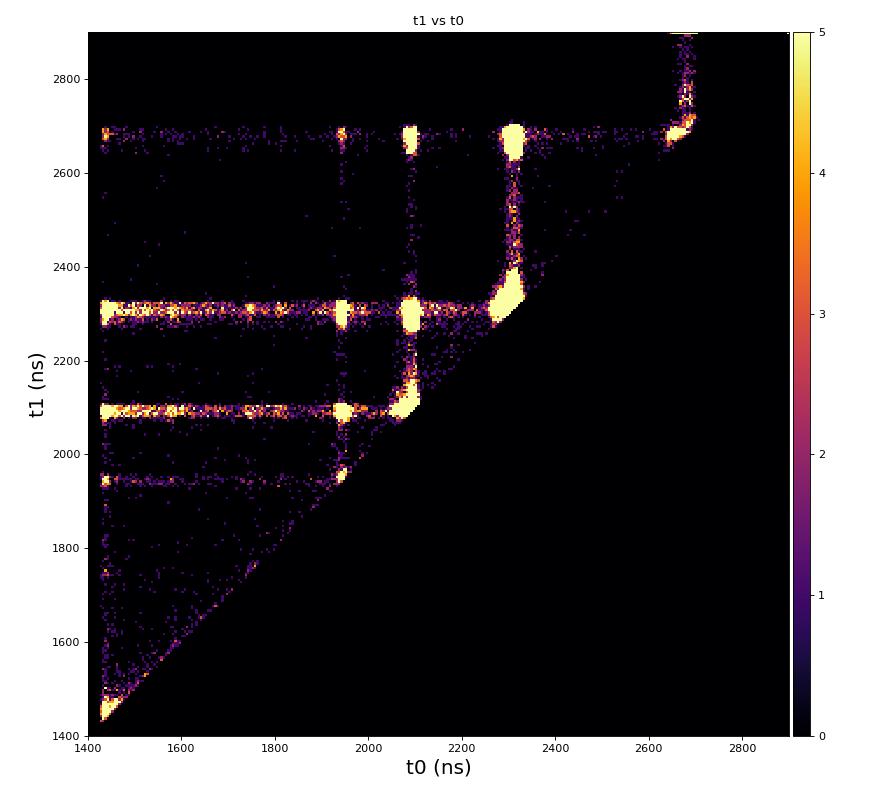
Only useful channels will be selected in xtc2. This hack is temporary available in
psana/psana/detector/test_detectors.py
Few days ago Chris asked me to check if RoentDec library interface still works. Due to relocation of different python and c++ modules in development of lcls2 software, compilation of examples for quad-anode was broken in ps-4.5.5/10. Now all visible issues are fixed and examples are working properly. Need in new release greater than ps-4.5.10.
Examples resides in lcls2/psana/psana/hexanode/examples/*
modified: ex-20-data-acqiris-access.py
modified: ex-21-data-acqiris-graph.py
modified: ex-22-data-acqiris-peaks-save-h5-xiangli.py
modified: ex-22-data-acqiris-peaks-save-h5.py
modified: ex-23-quad-proc-sort-graph-from-h5.py
modified: ex-24-quad-proc-sort-graph.py
modified: ex-25-quad-proc-data.py
modified: ex-26-calibconsts.py
modified: ex-27-calib-pop-rbfs-xiang.py
new file: ex_test_data.py
DIR_ROOT = os.getenv('DIR_PSDM') # /cds/group/psdm ON psana OR /sdf/group/lcls/ds/ana/ ON s3df
DIR_DATA_TEST = os.path.join(DIR_ROOT, 'detector/data2_test/xtc')
Then access to *.xtc2 files:
FNAME = '%s/%s' % (DIR_DATA_TEST, 'data-amox27716-r0100-acqiris-e000100.xtc2')
DIR_ABSPATH = os.path.abspath(os.path.dirname(__file__)) # absolute path to .../psana/hexanode/examples
+ 'calibcfg' : '%s/configuration_quad.txt' % DIR_ABSPATH,
+ 'calibtab' : '%s/calibration_table_data.txt' % DIR_ABSPATH,
All test examples are working.
References
Hexanode detector test on data
2019-11-01-Peter-Walter-article.pdf