Hi everyone,
Here is a short summary of what I heard today for how we should start
with the pnCCD.
For pnCCD algorithms:
common-mode, pedestals, hot-pixels, quadrant rotations, hit-finders,
support in mikhail's calibManager
For pnCCD online displays: (using matplotlib for now)
shot by shot raw data
shot by shot calibrated data
projections of the above
region-of-interest
strip-charts of interesting quantities
(also display calibration values like noise-map,pedestal-map)
After this we will work on the acqiris as well (acqiris
constant-fraction algos already exist in psana).
Attached below is a 12 line python program that plots a real pnCCD
image and an x-projection (amoc0113 also has pnccd data we can look
at). You can run it on a psana node by saving it to pnccd.py and
doing "sit_setup" and then "ipython pnccd.py". This sort of code
should work online too (although we may have to change matplotlib
settings) as well as with calibrated images.
Display group (dan, mikhail, me) meets Thursday at 10:30. Analysis
group (sebastian, ankush(?), phil, mikhail, me) meets Friday at 1.
See you then...
chris
Large area pnCCD DAQ and Elictronics, Lothar Struder & Robert Hartmann
Use interactive psana framework ~cpo/ipsana/shm.py:
from psana import *
events = DataSource('shmem=1_1_XCS.0').events()
src = Source('DetInfo(XcsBeamline.1:Tm6740.5)')
import matplotlib.pyplot as plt
plt.ion()
fig = plt.figure('pulnix')
ax = fig.add_axes([0.1, 0.1, 0.8, 0.8]) # x0, y0, h, w
for i in range(100):
evt = events.next()
frame = evt.get(Camera.FrameV1, src)
ax.cla()
ax.imshow(frame.data16())
fig.canvas.draw()
|


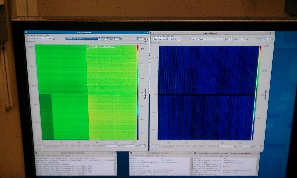
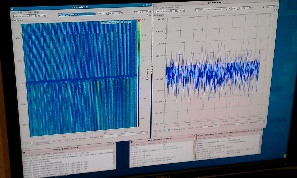
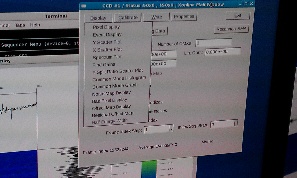
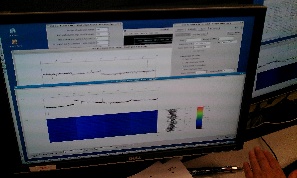
On 2014-01-27 Sebastian Carron kindly provide us with data files for pnCCD experiment amoa1214:
Example can be found in PSCalib/test/ex_calib_file_finder.cpp:
// Assume that file is located in /reg/d/psdm/AMO/amotut13/calib/PNCCD::CalibV1/Camp.0:pnCCD.1/pedestals/1-end.data #include "PSCalib/PnccdCalibPars.h" const std::string calib_dir = "/reg/d/psdm/AMO/amotut13/calib"; const std::string group = "PNCCD::CalibV1"; // or std::string() const std::string source = "Camp.0:pnCCD.1"; unsigned long runnum = 10; unsigned print_bits = 255; PSCalib::PnccdCalibPars *calibpars = new PSCalib::PnccdCalibPars(calib_dir, group, source, runnum, print_bits); calibpars->printCalibPars(); ndarray<CalibPars::pedestals_t, 3> peds = calibpars -> pedestals_ndarr(); ndarray<CalibPars::common_mode_t, 1> cmod = calibpars -> common_mode_ndarr(); ndarray<CalibPars::pixel_status_t, 3> stat = calibpars -> pixel_status_ndarr(); ndarray<CalibPars::pixel_gain_t, 3> gain = calibpars -> pixel_gain_ndarr(); ndarray<CalibPars::pixel_rms_t, 3> gain = calibpars -> pixel_gain_ndarr(); // OR: CalibPars::pedestals_t* p_peds = calibpars -> pedestals(); CalibPars::common_mode_t* p_cmod = calibpars -> common_mode(); CalibPars::pixel_status_t* p_stat = calibpars -> pixel_status(); CalibPars::pixel_gain_t* p_gain = calibpars -> pixel_gain(); CalibPars::pixel_rms_t* p_rms = calibpars -> pixel_rms(); const size_t ndim = ndim(); const size_t size = size(); const unsigned* shape = shape(); etc... |
973.941639 881.189675 1050.211 773.263749 899.241302 981.805836 1150.72615 993.084175 1121.15488 1029.76319 1220.14927 903.278339 1097.49944 1066.94949 1263.71044 1053.53872 1194.35915 935.320988 1317 ... |
pdscalibdata::PnccdPedestalsV1::pars_t = float
pdscalibdata::PnccdCommonModeV1::pars_t = uint16_t
pdscalibdata::PnccdPixelStatusV1::pars_t = uint16_t
pdscalibdata::PnccdPixelGainV1::pars_t = float
Factory is implemented for pnCCD only. CSPAD and CSPAD2x2 will be added soon. |
#include "PSCalib/CalibPars.h" #include "PSCalib/CalibParsStore.h" // Instatiation //Here we assume that code is working inside psana module where evt and env variables are defined through input parameters of call-back methods. //Code below instateates calibpars object using factory static method PSCalib::CalibParsStore::Create: std::string calib_dir = env.calibDir(); // or "/reg/d/psdm/<INS>/<experiment>/calib" std::string group = std::string(); // or something like "PNCCD::CalibV1"; const std::string source = "Camp.0:pnCCD.1"; const std::string key = ""; // key for raw data Pds::Src src; env.get(source, key, &src); PSCalib::CalibPars* calibpars = PSCalib::CalibParsStore::Create(calib_dir, group, src, PSCalib::getRunNumber(evt)); // Access methods calibpars->printCalibPars(); const PSCalib::CalibPars::pedestals_t* peds_data = calibpars->pedestals(); const PSCalib::CalibPars::pixel_gain_t* gain_data = calibpars->pixel_gain(); const PSCalib::CalibPars::pixel_rms_t* rms_data = calibpars->pixel_rms(); const PSCalib::CalibPars::pixel_status_t* stat_data = calibpars->pixel_status(); const PSCalib::CalibPars::common_mode_t* cmod_data = calibpars->common_mode(); |
In order to get rid of detector dependent types of calibration parameters we need to add metadata in the calibration file. All metadata can be listed in the header of the calibration files, for example, using keyward mapping (dictionary):
# RULES:
# Lines starting with # in the beginning of the file are considered as comments or pseudo-comments for metadata
# Lines without # with space-separated values are used for input of parameters
# Empty lines are ignored
# Optional fields:
# TITLE: This is a file with pedestals
# DATE_TIME: 2014-01-30 10:21:23
# AUTHOR: someone
# EXPERIMENT: amotut13
# DETECTOR: Camp.0:pnCCD.1
# CALIB_TYPE: pedestals
# Mandatory fields to define the ndarray<TYPE,NDIMS> and its shape as unsigned shape[NDIMS] = {DIM1,DIM2,DIM3}
# TYPE: float
# NDIMS: 3
# DIM1: 4
# DIM2: 255
# DIM3: 255
973.941639 881.189675 1050.211 773.263749 899.241302 981.805836 1150.72615 993.084175 1121.15488 1029.76319 1220.14927 903.278339 1097.49944 1066.94949 1263.71044 1053.53872 1194.35915 935.320988 1317 ... |
All other staff looks useful for all users and is migrated to the PSDM page: pnCCD processing pipeline