ImgMan User's Guide
- #Overview
- #Starting ImgMan from Matlab Desktop
- #Viewing Live Images
- #Collecting Images
- #Browsing Image Datasets
- #Analyzing Single Image
- #Help
Overview
ImgMan (short for Image Management) consists of the Image Acquisition, Image Browser, and Image Analysis windows.
Starting ImgMan from Matlab Desktop
To start ImgMan from Matlab, type
imgAcq_main
in the Command Window.
Viewing Live Images
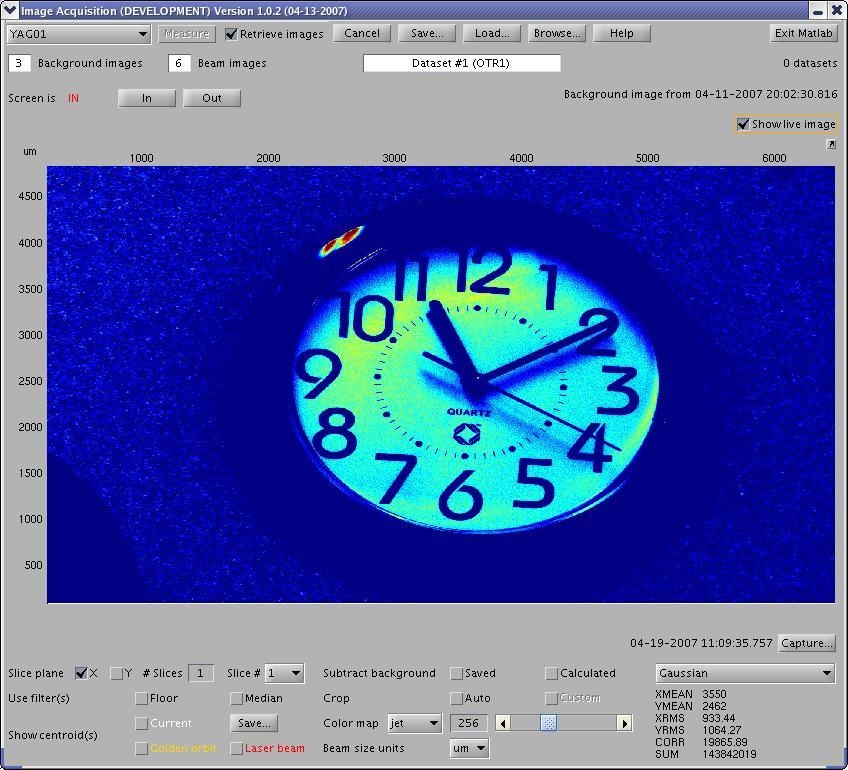
In the Image Acquistion window, select a camera and check the "Show Live Image" box. After a short delay, the window expands, and you can see the live image as well as the #Image Processing Panel, which can be found on all three main windows of ImgMan.
Image Processing Panel
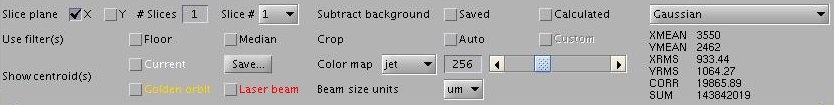
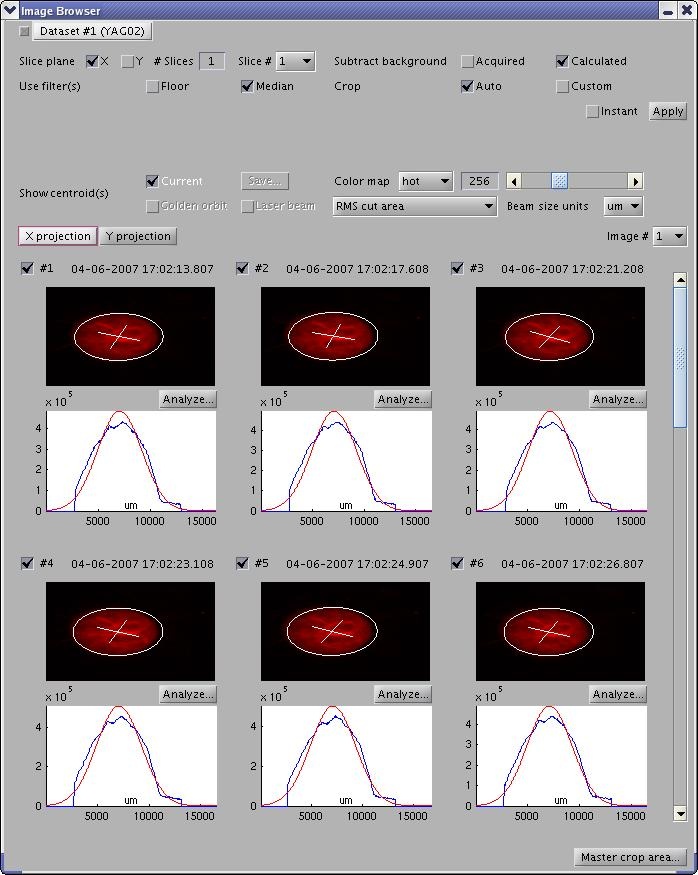
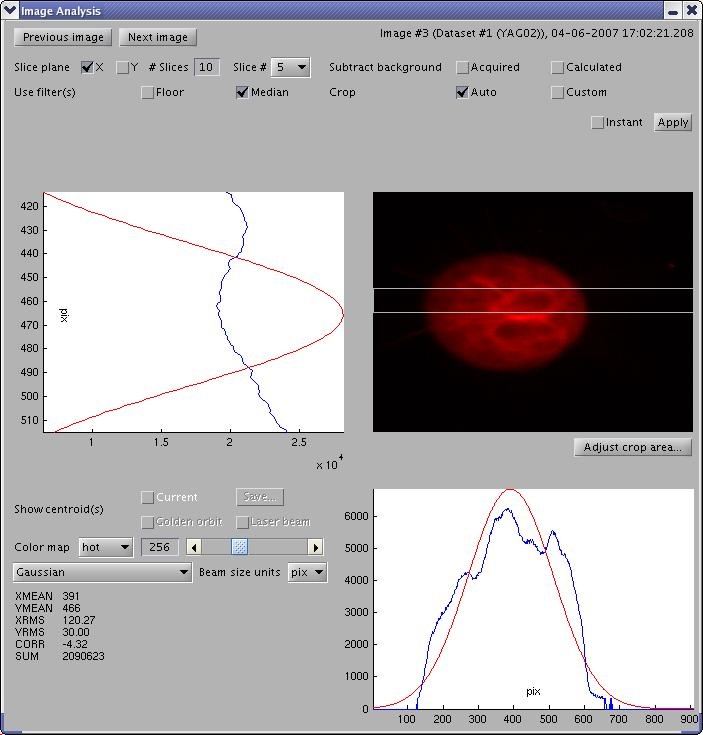
Image Processing Panel contains widgets for setting image processing parameters, e.g. slicing, background subtraction, filters, cropping, color mapps, annotations (centroids), unit scales, the image processing algorithm. Usually, ImgMan propagates changes of paramaters immediately. However, if there is an "Apply" button, you must press it before some costly image processing parameters are applied (or you can check the "Instant" Apply box).
Collecting Images
In the Image Acquisition window, select a camera, specify the desired number of background and beam images, and press the "Measure" button. If you don't want to save images locally, uncheck "Save Images" box. You can follow the progress of the measurment either in the Image Acquisition window or in cmlog. Each measurement results in one dataset of images. When the measurement is finished, the number on the dataset label increases by 1.
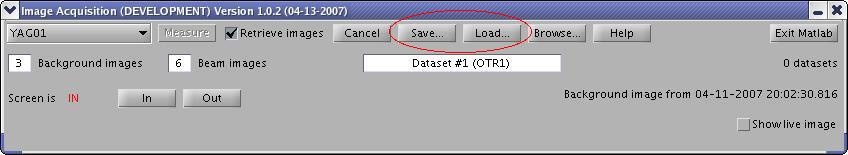
Saving and Loading Datasets
Press "Save" button to open "Save As..." dialog and save your image datasets. Press "Load" button to open a "Load..." dialog and load image datasets.
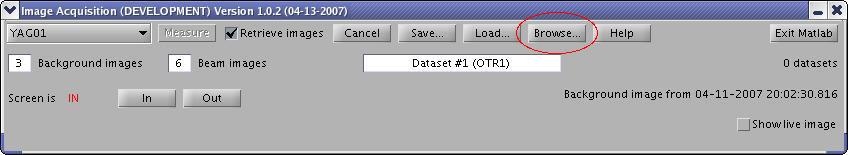
Browsing Image Datasets
In the Image Acquisition window, press the "Browse..." button to open the image browser. You can select a dataset, browse images, and specify image processing parameters through an #Image Processing Panel.
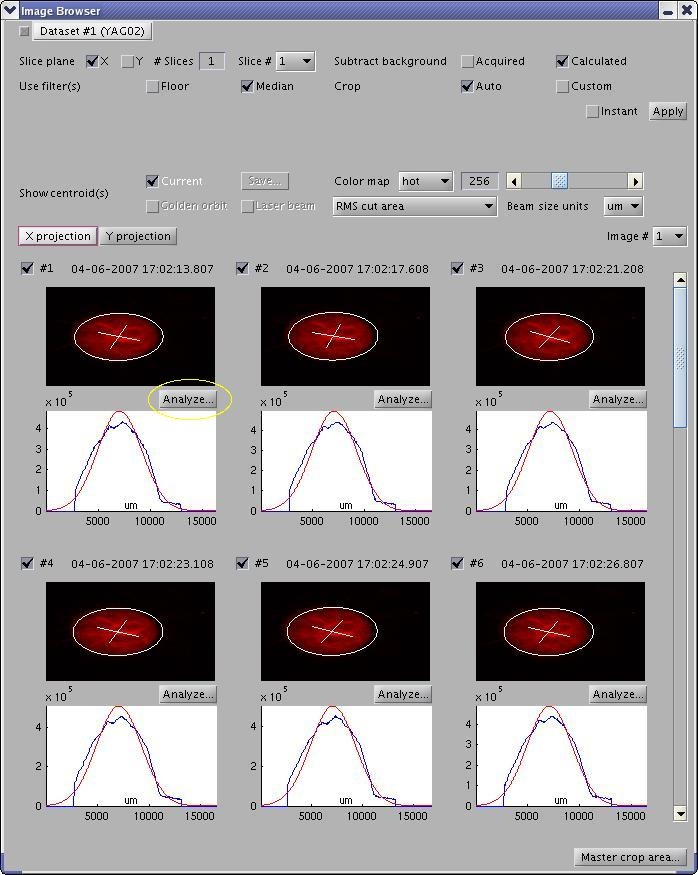
Analyzing Single Image
In the Image Browser window, press the "Analyze..." button to open an Image Analysis window for the corresponding image. Image Analysis also contains an #Image Processing Panel.
Help
Image Acquisition
ALGORITHM popupmenu
- Select an image processing algorithm.
BACKGROUND IMAGES edit
- Specify the number of background images to be acquired.
BEAM IMAGES edit
- Specify the number of beam images to be acquired.
BEAM SIZE UNITS popupmenu
- Select image axes units.
BROWSE... button
- Open the Image Browser.
CAMERA popupmenu
- Select a camera from the list.
CANCEL button
- Cancel the image acquisition.
CAPTURE... button
- Display the current live image (screenshot) in another window.
COLOR MAP edit
- Specify the number of colors.
COLOR MAP popupemu
- Specify the name of the colormap.
COLOR MAP slider
- Select the number of colors.
CROP checkboxes
- Select which cropping to apply to the image.
DATASET edit
- Specify the name of the next image dataset.
FILTERS checkboxes
- Select desired image filters.
HELP button
- Display help.
LOAD... button
- Load image datasets from hard drive.
MEASURE button
- Start image acquisition. Might be disabled.
NUMBER SLICES edit
- Specify the number of slices.
SAVE... button
- Save image datasets to hard drive.
SAVE.... CURRENT CENTROID button
- Open a dialog to save the current centroid.
SAVE IMAGES checkbox
- Enable/disable the local saving of images.
SHOW CENTROIDS checkboxes
- Select to display the desired centroids,
SLICE INDEX edit
- Specify the index of the slice.
SLICE PLANE checkboxes
- Select the plane for slicing.
SUBTRACT BACKGROUND checkboxes
- Select which background to subtract from the image.
UNDOCK button
- Display live image in another (resizable) figure.
Image Analysis
Image Browser