Restrictions
VASP is restricted to licensed users. It may not be used in contractual research in cooperation with industry or military.
Submitting Batch Jobs
vasp-ver-bsub <version> myscript.py vasp-ver-bsub-native <version> -q suncat-test -n 2 -o junk.log
See the bottom of this page for available versions.
The first is the ASE style. The second is native VASP style, and assumes all input files (POTCAR, INCAR, etc.) are created by the user are in the current directory. POTCARs can be assembled from the directory /nfs/slac/g/suncatfs/sw/vasp/pseudo52.
Example script
Thanks to Joel Varley for providing this BEEF example.
#!/usr/bin/env python
#LSF -q suncat-test
#LSF -n 2
#LSF -o std.out
#LSF -e std.err
from ase import *
from ase import io
from ase.lattice.spacegroup import crystal
from ase.io.trajectory import PickleTrajectory
from ase.optimize import BFGS, QuasiNewton
from ase.units import kJ, Hartree
from ase.utils.eos import EquationOfState
from ase.calculators.vasp import Vasp
from ase.data.molecules import molecule as molecule
from ase.vibrations import Vibrations
import os, sys
#############################################################################
#############################################################################
name = 'H'
a = 5.0
atoms = Atoms([Atom('H',(a/2, a/2, a/2))],
cell=(a, a, a))
# note that BEEF is only available in suncat vasp version 2 (v2) or later.
xcf = 'BEEF'
encut = 400
nkp = 1
sigma = 0.05
ibrion= -1 # no atomic displacement done by VASP (only ASE)
ispin = 1 # NOT spin-polarized (ispin=2 for SP)
# some types of GGA
vaspgga = {'RPBE' : 'RP',
'PBE' : 'PE',
'AM05' : 'AM',
'PBEsol' : 'PS',
'PW86' : 'PW',
'PW91' : '91',
'BEEF' : 'BF',
'GGA' : '--',
}
if xcf=='BEEF':
luse_vdw=True
if not (os.path.exists('vdw_kernel.bindat')):
os.symlink('/nfs/slac/g/suncatfs/sw/vasp/vdw_kernel.bindat','vdw_kernel.bindat')
else:
luse_vdw=False
# VASP uses LDA or PBE PAW pseudopotentials as a start (gga flag modifies the GGA paramaterization
if xcf in vaspgga.keys():
xc = 'PBE'
else:
xc = 'LDA'
vaspgga['LDA'] = '--'
#############################################################################
#############################################################################
calc = Vasp(encut=encut,
xc=xc,
gga=vaspgga[xcf],
kpts = (nkp,nkp,nkp),
gamma = False, # Gamma-centered (defaults to Monkhorst-Pack)
ismear=0,
sigma=sigma,
ibrion=ibrion,
nsw=0, # don't use the VASP internal relaxation, only use ASE
#potim=potim, # use to take smaller ionic steps (or for vibration/normal mode calculations)
lvtot=False,
npar=1, # use this if you run on one node (most calculations). see suncat confluence page for optimal setting
lwave=False,
luse_vdw=luse_vdw,
ispin=ispin,
zab_vdw=-1.8867, # this selects vdw-DF2
#lreal=True, # can speed up if supercells are large
)
atoms.set_calculator(calc)
dyn = QuasiNewton(atoms, logfile=name+'.log', trajectory=name+'.traj')
dyn.run(fmax=0.05)
electronicenergy = atoms.get_potential_energy()
f = open('out.energy','w')
f.write(str(electronicenergy))
f.close()
# clean up big leftover files (WAVECAR or CHGCAR could be of use for some calculations)
os.system('rm CHG vasprun.xml PCDAT WAVECAR CHGCAR')
#############################################################################
Versions
Version | Date | Comment |
1 | 12/5/2012 | initial version |
2,2a | 1/31/2013 | add BEEF support courtesy of Johannes Voss ("a" for suncat3 farm) |
3 | 1/31/2013 | like v2, but change pseudo potentials to the "52" versions. no suncat3 version yet |
4 | 2/28/2013 | like v3, but add VTST support |
5 | 3/15/2013 | includes BEEF, VTST, and 30% performance improvement from using fftmpiw.F |
6 | 7/9/2013 | copy of v5 but without -DNGZhalf flag. requested by junyan for a non-collinear calculation. not recommended for general use |
Parallelization
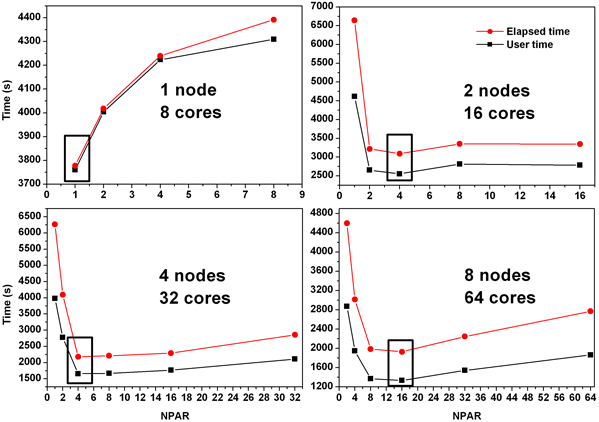
This information comes from Yian Zhu, who ran several 3x3x4 spin-polarized Nickel calculations with the PBE functional.
- VASP parallelizes over bands by default, but can also parallelize more efficiently over plane waves (controlled by the "NPAR" parameter, which by default is set to the total number of cores).
- #cores used per-band is totalNumberOfCores/NPAR
- Yian has measured how to set NPAR for the suncat 8-core farm for his 3x3x4 spin-polarized Nickel calculation. For one node NPAR should be set to 1 (planewave parallelization). For more nodes, NPAR should be set to approximately sqrt(numberOfCores), at least for Yian's system. See Yian's plot below.
- band parallelization requires less interprocess communication than planewave parallelization, but still requires non-trivial inter-node communication
- for Yian's system, even with an optimal setting of NPAR, he only sees an improvement from 3777 seconds to 3092 seconds going from 1 to 2 nodes, so it is still significantly more cpu-efficient to run on one node.