Motivation
Development of this application was stimulated by the discussion with Marcin Sikorski (meeting on 2012-08-30), doing xcs experiments.
Users need in real-time algorithm for calculation of image vs time auto-correlation function
g2(tau) = <I(t)*I(t+tau)> / (<I(t)> * <I(t+tau)>),
where I(t) is an image intensity at time t, and tau is a delay between two measurements.
Typical experimental condition can be described as follows:
- Run duration is about one hour at frequency up to 120 Hz that gives up to 10^5-10^6 images.
- Currently typical imaging devise is a Princeton camera with 1300x1340 pixels.
- Need to calculate
g2(tau)for each pixel, averaged over all possible image timestwith time differencetaubetween images. - A set of
taushould have about 30-100 points in log scale uniformly covering the run duration. - Use for example xcsi0112-r0015: 500 images with 8 sec delay between images.
Desired time for evaluation of the auto-correlation function should be comparable with run duration <1 hour. Currently this algorithm takes a few hours that can not be used for fast feedback in real time experiment.
Algorithm
Basic idea is (1) to split image vs time for small parts in image, (2) to process each part on separate computer node, (3) to merge results at the end of processing. It is clear that significant speedup (about T/N_nodes_) is achieved at the 2nd stage. These three stages are performed in separate C++ applications. Wrapping python script allows to submit job by a single command. It takes care about file and sub-process management in this job, as described below.
Code location
All modules for this application resides in the package ImgAlgos:
Module |
Functionality |
|---|---|
ImgVsTimeSplitInFiles |
splitter |
CorAna |
base class with common methods |
CorAnaData |
data processing for split files |
CorAnaInputParameters |
provides storage for input parameters |
CorAnaMergeFiles |
merging algorithm |
CorAnaProcResults |
Example showing how to access results using C++ and produce a table for presentation |
CorAnaPars.py |
singleton class for parameter storage in the wrapping file manager |
CorAnaSubmit.py |
global methods for the file manager |
app/corana_submit |
pythonic script which defines the sequence of procedures |
app/corana.cpp |
main module for the part of image vs time correlation processing |
app/corana_merge.cpp |
main module for merging |
app/corana_procres.cpp |
main module for processing of results from correlator array |
data/psana-corana.cfg |
psana configuration file for ImgVsTimeSplitInFiles |
data/PlotCorAnaResults.py |
example of the python script which plots the resulting graphics |
Image splitting
Image splitting is implemented as a regular psana module ImgAlgos::ImgVsTimeSplitInFiles.
Command to run interactively on psana#### or submit in batch from pslogin## node:
psana -c <config-file> <xtc-file-list> bsub -q psfehq -o log-file 'psana -c <config-file> <xtc-file-list>'
For example:
psana -c ImgAlgos/data/psana-corana.cfg /reg/d/psdm/XCS/xcsi0112/xtc/e167-r0015-*
where ImgAlgos/data/psana-corana.cfg is an example of the configuration script for psana and /reg/d/psdm/XCS/xcsi0112/xtc/e167-r0015-* are the input xtc files for particular run.
A couple of limitations due to LCLS policy:
Interactive job can be run on psana#### computer, but the batch queues are not seen from psana#### nodes...
Batch job can be submitted from pslogin## computer, but data are not seen directly from pslogin## nodes...
Produces the files:
cor-ana-r0015-b0000.bin - file with a part of image vs time cor-ana-r0015-b0001.bin cor-ana-r0015-b0002.bin cor-ana-r0015-b0003.bin cor-ana-r0015-b0004.bin cor-ana-r0015-b0005.bin cor-ana-r0015-b0006.bin cor-ana-r0015-b0007.bin cor-ana-r0015-time.txt - list of time-records for all events in processed run. cor-ana-r0015-time-ind.txt - list of time-records for all events in processed run with time index. cor-ana-r0015-med.txt - file with metadata. In particular it has the original image size, number of image parts for splitting, number of images in run, etc.
Algorithms:
- The <int16_t> image data array is split for ordered number of equal parts (by the parameters
nfiles_outin psana-corana.cfg file) and each part is saved in the outputcor-ana-r0015-b####.binfile sequentially for all selected events. - The appropriate time record for selected event is saved in the file
cor-ana-r0015-time.txt. - At the end of the splitting procedure:
- the average time difference and its rms between sequential events is evaluated for all recorded time records.
- The file
cor-ana-r0015-time.txtis re-processed and for each record the time index is evaluated as unsigned value of<time-index> = (<event-time> + 0.5 <average-time-between-events>) / <average-time-between-events>
- Event record with time index is saved in the file
cor-ana-r0015-time-ind.txt
- All metadata parameters which are required for further processing, such as input parameters, image size,
<average-time-between-events, maximal value of the time index etc., are saved in filecor-ana-r0015-med.txt.
This approach allows to apply the modest event selection algorithms in psana pre-processing stage.
But, it still based on uniform time indexing...
Q: Is it really good assumption for this kind of experiments?
Time correlation processing
ImgAlgos/app/corana application
Command to run interactively on psana#### or submit in batch from pslogin## node:
corana -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h] bsub -q psfehq -o log-file 'corana -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]'
For example the interactive and batch mode commands:
corana -f cor-ana-r0015-b0001.bin -t my-tau.txt bsub -q psfehq -o log-file 'corana -f cor-ana-r0015-b0000.bin'
Produce files:
cor-ana-r0015-tau.txt - string of {{tau}} values for which the auto-correlation function is evaluated
cor-ana-r0015-b0000-result.bin - auto-correlators for the part of the image for all {{tau}} values
cor-ana-r0015-b0001-result.bin
cor-ana-r0015-b0002-result.bin
cor-ana-r0015-b0003-result.bin
cor-ana-r0015-b0004-result.bin
cor-ana-r0015-b0005-result.bin
cor-ana-r0015-b0006-result.bin
cor-ana-r0015-b0007-result.bin
Merging results
ImgAlgos/app/corana_merge application
Command to run interactively on psana#### or submit in batch from pslogin## node:
corana_merge -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h] bsub -q psfehq -o log-file 'corana_merge -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]'
For example:
corana_merge -f cor-ana-r0015-b0001-result.bin -t my-tau.txt
This procedure produces file:
cor-ana-r0015-image-result.bin
Example of how to get and process results
ImgAlgos/app/corana_procres
Command to run interactively on psana#### or submit in batch from pslogin## node:
corana_procres -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h] bsub -q psfehq -o log-file 'corana_procres -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]'
Basically it reads files with results and produces the histogram-like table *-hist.txt.
Automatic processing
ImgAlgos/app/corana_submit - is a wrapping script which allows to run all of above procedures by a single command from pslogin## node and it keeps eye on processing of jobs in batch and doing the file management. Command to start:
corana_submit [-c <config-file>] [-t <fname-tau>] [-x] <xtc-file-list>
For example:
corana_submit -c ImgAlgos/data/psana-corana.cfg -t my-tau.txt /reg/d/psdm/XCS/xcsi0112/xtc/e167-r0015-s00-c00.xtc
This script sequentially performs operations for single run as follows:
- Initialize all parameters
- Run psana to split image for files
- Check that all split files are produced
- Submit job for time-correlation processing
- Check that all processed files are produced
- Submit job for merging
- Check that merged file is produced
- Submit job for test processing of the file with results
- List all created files
- Clean-up files in the work directory
- List of preserved files
The next to last procedure deletes all intermediate split- and log- files.
In debugging mode this procedure may be turned off.
Manual sequential processing
In case of manual processing of all scripts, commands need to be issued in a right order. Commands corana, corana_merge, and corana_procres should have the same list of parameters. This is important, because all file names for these procedures are generated by the same base class ImgAlgos/src/CorAna.cpp
Right sequence of commands to run interactively on psana####
psana -c <config-file> <xtc-file-list> corana -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h] corana_merge -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h] corana_procres -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]
or submit in batch from pslogin## node:
bsub -q psfehq -o log-file 'psana -c <config-file> <xtc-file-list>' bsub -q psfehq -o log-file 'corana -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]' bsub -q psfehq -o log-file 'corana_merge -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]' bsub -q psfehq -o log-file 'corana_procres -f <fname-data> [-t <fname-tau>] [-l <logfile>] [-h]'
The corana batch jobs can be submitted and run on separate butch nodes in parallel. All other procedures can be submitted when previous is successfully finished and all necessary files are produced.
The corana_procres command is optional and is currently used for test purpose only. But, it may be replaced by real analysis code.
File formats
- File with split-image data for selected events
cor-ana-r0015-b000N.bin:
Currently this file contains<uint16_t>amplitude for each pixel in binary format for:<data-for-img-partN-of-img1> <data-for-img-partN-of-img2> ... <data-for-img-partN-of-imgLast>
- File with metadata parameters
cor-ana-r0015-med.txt:IMAGE_ROWS 1300 IMAGE_COLS 1340 IMAGE_SIZE 1742000 NUMBER_OF_FILES 8 BLOCK_SIZE 217750 REST_SIZE 0 NUMBER_OF_IMGS 500 FILE_TYPE bin DATA_TYPE uint16_t TIME_SEC_AVE 8.088413 TIME_SEC_RMS 0.063639 TIME_INDEX_MAX 499
- File with image time records
cor-ana-r0015-time.txt:where each record has:1 0.000000 0.000000 20120616-080236.671607864 5366 0 2 8.026429 8.026429 20120616-080244.698036743 8255 1 3 16.144788 8.118359 20120616-080252.816395836 11177 2 4 24.154835 8.010048 20120616-080300.826443448 14060 3 ...<image-in-file#> <t(sec)-from-the-1st-event> <dt(sec)> <time-stamp> <fiducials> <event#-since-configure>
- File with image time records and evaluated time index
cor-ana-r0015-time-ind.txt:where each record has:1 0.000000 0.000000 20120616-080236.671607864 5366 0 0 2 8.026429 8.026429 20120616-080244.698036743 8255 1 1 3 16.144788 8.118359 20120616-080252.816395836 11177 2 2 4 24.154835 8.010048 20120616-080300.826443448 14060 3 3 5 32.281937 8.127102 20120616-080308.953545010 16985 4 4 ...<image-in-file#> <t(sec)-from-the-1st-event> <dt(sec)> <time-stamp> <fiducials> <event#-since-configure> <time-index-starting-from-0>
- File with split-image correlators for each value of
taucor-ana-r0015-b000N-result.bin:
Currently it saves<float>correlator for each pixel in binary format for:<corr-for-img-partN-of-tau1> <corr-for-img-partN-of-tau2> ... <corr-for-img-partN-of-tauLast>
my-tau.txt:contains the1 3 5 7 9 10 12 14 16 18 20 24 28 30 32 36 40 ... 160 180 200 240 280 300 320 360 400
tauvalues presented in terms of number of ordered images in the file.
Quick start guide
We assume that everything is set up to work on LCLS analysis farm, otherwise see Computing (including Analysis) and Account Setup.
How to run this procedure
If the version of the package ImgAlgos is available as a current software release, then you may run the script command(s) directly, for example:
cd <your-favorite-directory> mkdir work_corana sit_setup corana_submit [-c <config-file>] [-t <fname-tau>] [-x] <xtc-file-list>
If the code in the package ImgAlgos has been recently changed and the updated release is not yet available, then one need to create the local release directory, get the latest/HEAD version of the package, and compile the code as shown below:
cd <your-favorite-directory> newrel ana-current myReleaseDirectory cd myReleaseDirectory sit_setup addpkg ImgAlgos HEAD scons
Where to find results
The procedure will produce a bunch of files in the work_corana directory. If everything is OK, then all spit - and log- files will be removed at the end of automatic corana_submit procedure. The most important files are preserved for further analysis:
File name tail |
Format |
Content |
|---|---|---|
*-image-result.bin |
binary for <float> |
correlators for all image pixels for all tau values |
*-time-ind.txt |
text |
time records for all selected events/images |
*-tau.txt |
text |
the list of tau intervals |
*-med.txt |
text |
meta data parameters |
*-hist.txt |
text |
Histogram array with correlators averaged for ring regions of the image for all |
How to look at results
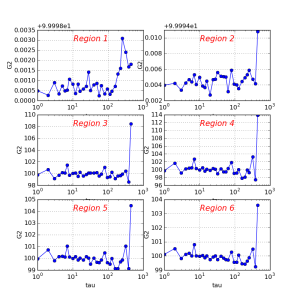
It is assumed that all files listed in previous section may be used for further analysis, depending on particular goals. The optional script corana_procres is designed as an example of how to access data from C++ code. Class CorAnaProcResults produces the file *-hist.txt
A simple python script shows how to plot this file:
./ImgAlgos/data/PlotCorAnaResults.py work_corana/cor-ana-r0015-hist.txt
Another option is to use python script for direct processing of the resulting files.
This is not elaborated yet.
Q: What kind of further processing is desired and what tools are going to be used?