Back to CSPAD Alignment
Content
Optical measurements
Comments from Gabriel Blaj:
Metrology file for CXI-Camera2-V16-20160205, replacing CXI-Camera2-V16-20150120, for all experiments taking place on or after February 05, 2016.
The only changes from the previous version are Quads 0 and 1.
Metrology files:
Quality check in X-Y plane
segm: S1 S2 dS1 dS2 L1 L2 dL1 dL2 angle(deg) D1 D2 dD d(dS) d(dL)
Quad 0
segm: 0 -20912 -20948 27 -9 43547 43552 -4 1 0.01184 48312 48323 -11 36 -5
segm: 1 -20879 -20913 115 81 43550 43548 36 34 0.12893 48292 48313 -21 34 2
segm: 2 -20917 -20919 -47 -49 -43551 -43547 24 28 -0.06315 48309 48314 -5 2 -4
segm: 3 -20959 -20925 -150 -116 -43550 -43549 52 53 -0.17498 48333 48313 20 -34 -1
segm: 4 20910 21118 -99 109 -43545 -43547 -48 -50 -0.00658 48397 48305 92 -208 2
segm: 5 20920 20907 -8 -21 -43556 -43566 -11 -21 0.01907 48329 48314 15 13 10
segm: 6 -20941 -20940 -39 -38 -43574 -43510 -44 20 -0.05066 48343 48288 55 -1 -64
segm: 7 -20915 -21020 97 -8 -43562 -43549 -3 10 0.05854 48317 48362 -45 105 -13
Quad 1
segm: 0 -20916 -20912 -109 -105 43549 43548 -48 -49 -0.14078 48312 48307 5 -4 1
segm: 1 -20916 -20917 -6 -7 43547 43225 -5 -327 -0.00858 48018 48311 -293 1 322
segm: 2 -20936 -20927 31 40 -43568 -43541 -30 -3 0.04670 48322 48324 -2 -9 -27
segm: 3 -20950 -20919 68 99 -43608 -43545 -74 -11 0.10979 48346 48342 4 -31 -63
segm: 4 20965 20753 19 -193 -43560 -43549 -10 1 0.11445 48258 48325 -67 212 -11
segm: 5 20934 20913 33 12 -43551 -43570 32 13 -0.02959 48314 48335 -21 21 19
segm: 6 -20927 -20919 95 103 -43570 -43578 -42 -50 0.13018 48335 48338 -3 -8 8
segm: 7 -20941 -21387 386 -60 -43570 -43547 20 43 0.21441 48328 48527 -199 446 -23
This check for original metrology file shows a few problems with
- Quad 0, segments 4 and 7
- Quad 1, segments 1, 4, and 7
Corrections
Quad 0:
- segment 4: point 18 Y: 67674 -> 67466
- segment 7: point 28 X: 65321 -> 65216
Quad 1:
- segment 1: point 4 X: 43224 -> 43546
- segment 4: point 18 Y: 67604 -> 67816
- segment 7: point 28 Y: 65798 -> 65352
Quality check in X-Y plane for corrected data
segm: S1 S2 dS1 dS2 L1 L2 dL1 dL2 angle(deg) D1 D2 dD d(dS) d(dL)
Quad 0
segm: 0 -20912 -20948 27 -9 43547 43552 -4 1 0.01184 48312 48323 -11 36 -5
segm: 1 -20879 -20913 115 81 43550 43548 36 34 0.12893 48292 48313 -21 34 2
segm: 2 -20917 -20919 -47 -49 -43551 -43547 24 28 -0.06315 48309 48314 -5 2 -4
segm: 3 -20959 -20925 -150 -116 -43550 -43549 52 53 -0.17498 48333 48313 20 -34 -1
segm: 4 20910 20910 -99 -99 -43545 -43547 -48 -50 0.13026 48307 48305 2 0 2
segm: 5 20920 20907 -8 -21 -43556 -43566 -11 -21 0.01907 48329 48314 15 13 10
segm: 6 -20941 -20940 -39 -38 -43574 -43510 -44 20 -0.05066 48343 48288 55 -1 -64
segm: 7 -20915 -20915 -8 -8 -43562 -43549 -3 10 -0.01052 48317 48316 1 0 -13
Quad 1
segm: 0 -20916 -20912 -109 -105 43549 43548 -48 -49 -0.14078 48312 48307 5 -4 1
segm: 1 -20916 -20917 -6 -7 43547 43547 -5 -5 -0.00855 48308 48311 -3 1 0
segm: 2 -20936 -20927 31 40 -43568 -43541 -30 -3 0.04670 48322 48324 -2 -9 -27
segm: 3 -20950 -20919 68 99 -43608 -43545 -74 -11 0.10979 48346 48342 4 -31 -63
segm: 4 20965 20965 19 19 -43560 -43549 -10 1 -0.02499 48349 48325 24 0 -11
segm: 5 20934 20913 33 12 -43551 -43570 32 13 -0.02959 48314 48335 -21 21 19
segm: 6 -20927 -20919 95 103 -43570 -43578 -42 -50 0.13018 48335 48338 -3 -8 8
segm: 7 -20941 -20941 -60 -60 -43570 -43547 20 43 -0.07892 48328 48333 -5 0 -23
Quad 2
segm: 0 -20944 -20953 91 82 43586 43585 40 39 0.11371 48356 48360 -4 9 1
segm: 1 -20973 -20937 -121 -85 43574 43547 -53 -80 -0.13548 48323 48354 -31 -36 27
segm: 2 -20911 -20953 10 -32 -43566 -43572 -7 -13 -0.01447 48350 48322 28 42 6
segm: 3 -20908 -20909 -52 -53 -43544 -43546 27 25 -0.06908 48303 48305 -2 1 2
segm: 4 20947 20911 58 22 -43547 -43547 9 9 -0.05263 48324 48306 18 36 0
segm: 5 20917 20911 -86 -92 -43548 -43554 -38 -44 0.11709 48310 48314 -4 6 6
segm: 6 -20908 -20907 66 67 -43545 -43572 -26 -53 0.08747 48323 48309 14 -1 27
segm: 7 -20913 -20909 -46 -42 -43483 -43480 23 26 -0.05798 48245 48251 -6 -4 -3
Quad 3
segm: 0 -21001 -20956 83 128 43541 43568 63 90 0.13878 48366 48320 46 -45 -27
segm: 1 -20906 -20904 0 2 43562 43543 19 0 0.00132 48318 48301 17 -2 19
segm: 2 -20932 -20909 -38 -15 -43544 -43576 22 -10 -0.03486 48329 48317 12 -23 32
segm: 3 -20934 -20904 -158 -128 -43550 -43552 63 61 -0.18813 48320 48308 12 -30 2
segm: 4 20912 20949 -69 -32 -43545 -43549 -44 -48 0.06644 48335 48297 38 -37 4
segm: 5 20938 20934 -157 -161 -43558 -43544 -87 -73 0.20918 48325 48319 6 4 -14
segm: 6 -20913 -20911 -150 -148 -43544 -43546 72 70 -0.19605 48306 48305 1 -2 2
segm: 7 -20921 -20918 -78 -75 -43541 -43552 39 28 -0.10065 48313 48307 6 -3 11
Quality check in Z
segm: SA LA XSize YSize dZS1 dZS2 dZL1 dZL2 dZSA dZLA ddZS ddZL dZX dZY angXZ(deg) angYZ(deg) dz3(um)
Quad 0
segm: 0 -20930 43549 43549 20930 -64 -10 -100 -46 -37 -73 -54 -54 -73 -37 -0.09604 -0.10129 54.115
segm: 1 -20896 43549 43549 20896 -8 51 -55 4 21 -25 -59 -59 -25 21 -0.03289 0.05758 59.013
segm: 2 -20918 -43549 20918 43549 -25 -4 -15 6 -14 -4 -21 -21 -14 -4 -0.03835 -0.00526 -21.001
segm: 3 -20942 -43549 20942 43549 50 30 48 28 40 38 20 20 40 38 0.10944 0.05000 19.951
segm: 4 20910 -43546 43546 20910 28 119 -76 15 73 -30 -91 -91 -30 73 -0.03947 0.20003 91.002
segm: 5 20913 -43561 43561 20913 33 406 -351 22 219 -164 -373 -373 -164 219 -0.21571 0.59998 373.251
segm: 6 -20940 -43542 20940 43542 51 53 -125 -123 52 -124 -2 -2 52 -124 0.14228 -0.16317 -1.821
segm: 7 -20915 -43555 20915 43555 24 37 -39 -26 30 -32 -13 -13 30 -32 0.08218 -0.04210 -12.992
Quad 1
segm: 0 -20914 43548 43548 20914 -116 41 -41 116 -37 37 -157 -157 37 -37 0.04868 -0.10136 156.978
segm: 1 -20916 43547 43547 20916 23 48 75 100 35 87 -25 -25 87 35 0.11447 0.09588 24.999
segm: 2 -20931 -43554 20931 43554 -24 -9 172 187 -16 179 -15 -15 -16 179 -0.04380 0.23548 -15.112
segm: 3 -20934 -43576 20934 43576 -12 -18 223 217 -15 220 6 6 -15 220 -0.04105 0.28926 5.713
segm: 4 20965 -43554 43554 20965 -60 -43 -45 -28 -51 -36 -17 -17 -36 -51 -0.04736 -0.13938 16.989
segm: 5 20923 -43560 43560 20923 -29 69 -117 -19 20 -68 -98 -98 -68 20 -0.08944 0.05477 98.119
segm: 6 -20923 -43574 20923 43574 42 19 56 33 30 44 23 23 30 44 0.08215 0.05786 22.999
segm: 7 -20941 -43558 20941 43558 -54 -21 19 52 -37 35 -33 -33 -37 35 -0.10123 0.04604 -33.027
Quad 2
segm: 0 -20948 43585 43585 20948 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 1 -20955 43560 43560 20955 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 2 -20932 -43569 20932 43569 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 3 -20908 -43545 20908 43545 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 4 20929 -43547 43547 20929 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 5 20914 -43551 43551 20914 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 6 -20907 -43558 20907 43558 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
segm: 7 -20911 -43481 20911 43481 0 0 0 0 0 0 0 0 0 0 0.00000 0.00000 0.000
Quad 3
segm: 0 -20978 43554 43554 20978 76 -1 -48 -125 37 -86 77 77 -86 37 -0.11313 0.10106 -76.759
segm: 1 -20905 43552 43552 20905 -27 -21 -48 -42 -24 -45 -6 -6 -45 -24 -0.05920 -0.06578 5.979
segm: 2 -20920 -43560 20920 43560 22 25 -30 -27 23 -28 -3 -3 23 -28 0.06299 -0.03683 -3.047
segm: 3 -20919 -43551 20919 43551 26 5 -31 -52 15 -41 21 21 15 -41 0.04108 -0.05394 20.991
segm: 4 20930 -43547 43547 20930 63 239 -265 -89 151 -177 -176 -176 -177 151 -0.23288 0.41335 175.587
segm: 5 20936 -43551 43551 20936 56 59 -61 -58 57 -59 -3 -3 -59 57 -0.07762 0.15599 2.992
segm: 6 -20912 -43545 20912 43545 23 15 -68 -76 19 -72 8 8 19 -72 0.05206 -0.09474 7.995
segm: 7 -20919 -43546 20919 43546 7 -5 -48 -60 1 -54 12 12 1 -54 0.00274 -0.07105 11.986
Constants
 Archive: /reg/g/psdm/detector/alignment/cspad/calib-cxi-camera2-2016-02-05/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data
Archive: /reg/g/psdm/detector/alignment/cspad/calib-cxi-camera2-2016-02-05/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data
 first deployed as /reg/d/psdm/CXI/cxi01516/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data
first deployed as /reg/d/psdm/CXI/cxi01516/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data
Quads alignment
2016-02-18 Alignment using Ag-behenate
Use Ag-behenate data of cxi01516 runs 21-23
Start with new optical measurements in /reg/g/psdm/detector/alignment/cspad/calib-cxi-camera2-2016-02-05/
Commands:
# get averaged data
det_ndarr_average -d exp=cxi01516:run=21 -s CxiDs2.0:Cspad.0 -f nda-cspad-Ag-behenate -n 10000 -p -v
# tune geometry
geo -g geo-cxi01516-2016-02-05.data -i nda-cspad-cxi01516-r0022-e010000-CxiDs2-0-Cspad-0-ave-Ag-behenate.txt -x 1000 -y 1000 &
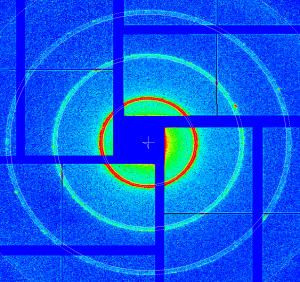
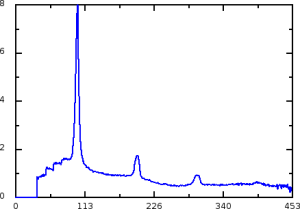
Archive: calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/geo-cxi01516-2016-02-18-Ag-behenate-tuned.data
Essential constants:
CSPAD:V1 0 QUAD:V1 0 -5100 -3000 0 90.0 0.0 0.0 0.00000 0.00000 0.00000
CSPAD:V1 0 QUAD:V1 1 -2800 6350 0 0.0 0.0 0.0 0.00000 0.00000 0.00000
CSPAD:V1 0 QUAD:V1 2 6350 4800 0 270.0 0.0 0.0 0.00000 0.00000 0.00000
CSPAD:V1 0 QUAD:V1 3 4800 -4900 0 180.0 0.0 0.0 0.00000 0.00000 0.00000
RAIL 0 CSPAD:V1 0 -1300 -3000 1000000 0.0 0.0 0.0 0.00000 0.00000 0.00000
IP 0 RAIL 0 0 0 0 0.0 0.0 0.0 0.00000 0.00000 0.00000
2016-03-11 Alignment using combination of runs
All files are located under directory: /reg/g/psdm/detector/alignment/cspad/calib-cxi-camera2-2016-02-05
Stage 1
Use very good Ag-behenate data cxi01516-r0022 and align all quads using this sample.
geo -x1000 -y1000 -g geo-cxi01516-2016-02-18.data -i nda-Ag-behenate-Z200-cxi01516-r0022-e012320-CxiDs2-0-Cspad-0-max.txt- create geometry file:
geo-cxi01516-r0022-2016-03-11.txt - images:
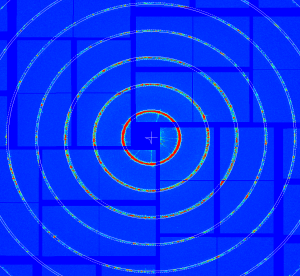
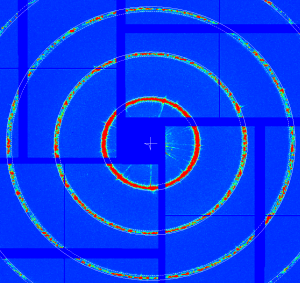
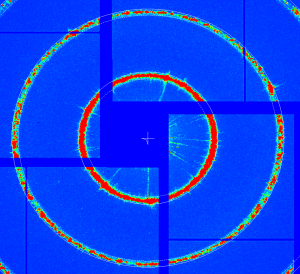
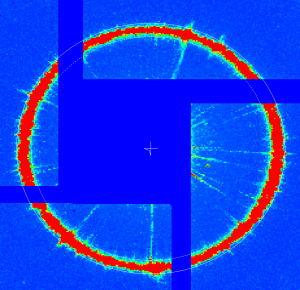
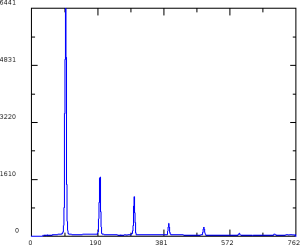
At further stages only minor (≤ 1pixel) quad corrections were applied
Stage 2
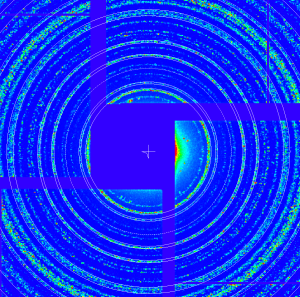
Use bright lysozyme data from run cxi01516-r0026
geo -x1000 -y1000 -g geo-cxi01516-r0022-2016-03-11.txt -i nda-lysozyme-cxi01516-r0026-e093480-CxiDs2-0-Cspad-0-max.txt- move entire detector, create geometry file
geo-cxi01516-r0026-2016-03-11.txt - images:
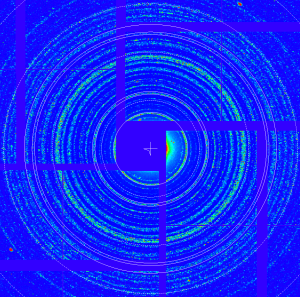
Stage 3
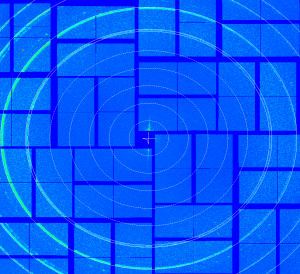
Use nozzle-ring data from run cxi02416-r0008
geo -x1000 -y1000 -g geo-cxi01516-r0026-2016-03-11.txt -i nda-nozzle-ring-cxi02416-r0008-e069865-CxiDs2-0-Cspad-0-max.txt- move entire detector, create geometry file
geo-cxi02416-r0008-2016-03-11.txt - image:
Stage 4
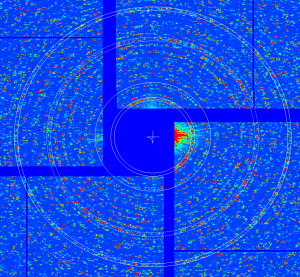
Use barely seen lysozyme data from run me-cxi02416-r0010
-
geo -x1000 -y1000 -g geo-cxi02416-r0008-2016-03-11.txt -i nda-lysozyme-cxi02416-r0010-e052421-CxiDs2-0-Cspad-0-max.txt - move entire detector, create geometry file
geo-cxi02416-r0010-2016-03-11.txt - image:
Alignment conclusion:
- Quad alignment obtained with
nda-Ag-behenate run cxi01516-r0022 works fine with all other data - For each other data-set, if detector was moved its position needs to be re-aligned.
Further cross-check
- use
geo-cxi02416-r0010-2016-03-11.txt and apply Oleksandr's geometry correction - check corrected geometry with cxi01516 run22 and 26
References
Archive: /reg/g/psdm/detector/alignment/cspad/calib-cxi-camera2-2016-02-05/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data
first deployed as /reg/d/psdm/CXI/cxi01516/calib/CsPad::CalibV1/CxiDs2.0:Cspad.0/geometry/1-end.data










