Table of Contents
Useful links
CP groups
Technical
- SDF guide and documentation, particularly on using Jupyter notebooks interactively or through web interface (runs on top of nodes managed by SLURM)
- Training dataset dumper (used for producing h5 files from FTAG derivations) documentation and git (Prajita's fork, bjet_regression is the main branch)
- SALT documentation, SALT on SDF, puma git repo (used for plotting), and Umami docs (for postprocessing), also umami-preprocessing (UPP)
- SLAC GitLab group for plotting related code
export PATH="/sdf/group/atlas/sw/conda/bin:$PATH"
FTAG1 derivation definition (FTAG1.py)
Documents and notes
- GN1 June 2022 PUB note, nice slides and proceedings from A. Duperrin
- Jannicke's thesis (chapter 4 on b-jets)
Presentations and meetings
- See all B-jet calibration meetings on Indico
- Framework experience (Prajita, July 6)
- Plans (Prajita, July 13)
- What needs to be added to JETM2 (August 17)
- Talk at Higgs workshop (October 4)
- Update from Brendon on, first training with large dataset and reasonable performance (October 23)
- New JETMX derivation formats (JETM2 on S10-13)
Samples
- PMG central page (MC20)
- JETM2 non-allhadronic ttbar mc20 (JIRA)
- MC20, MC21 & MC23 Sample List from Elisabeth for different physics processes
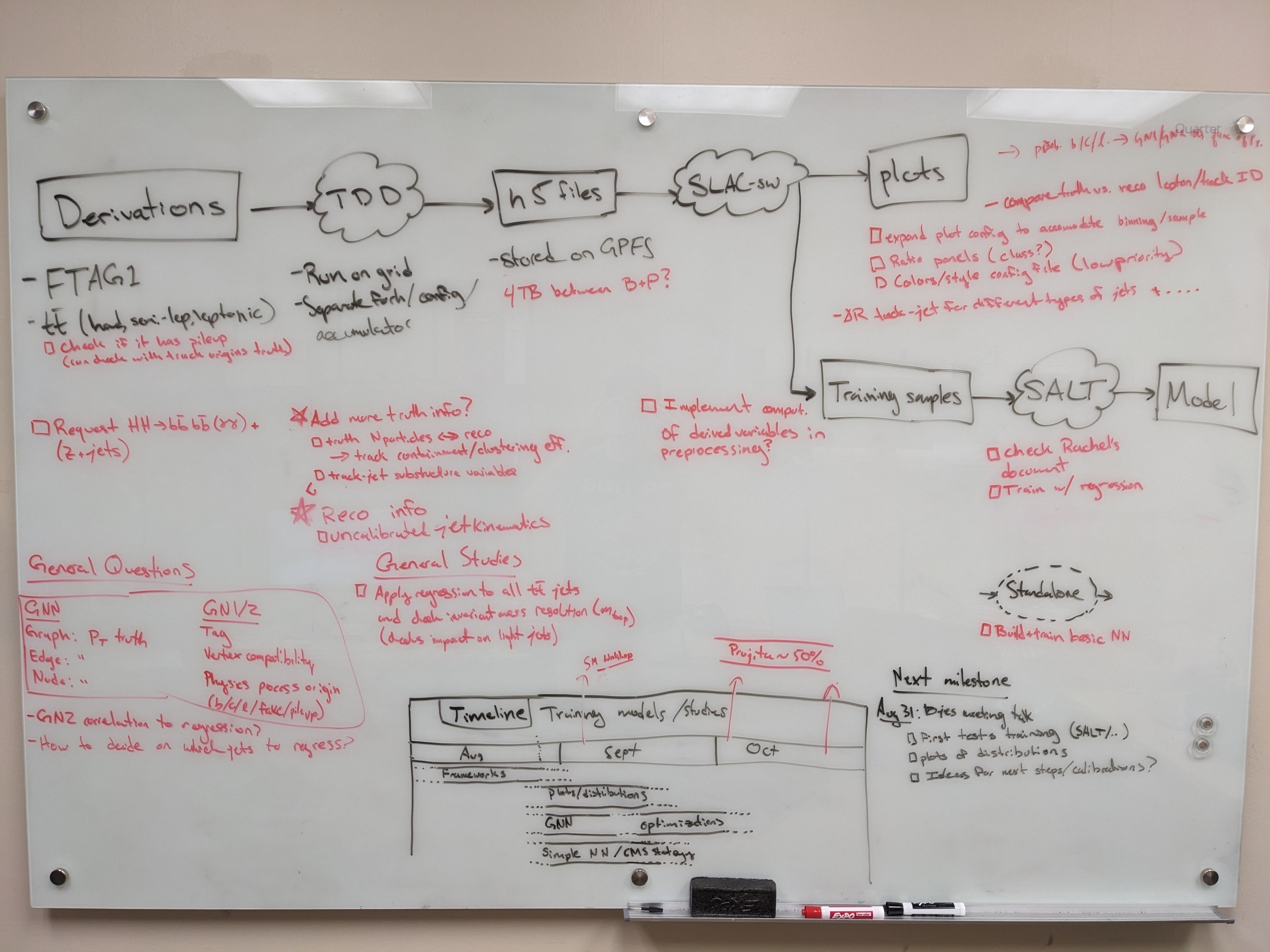
Planning
Ongoing studies
- Understanding bias in JES in low truth-jet pT
- Truth-jet matching (Brendon)
- Check different truth jet matching schemes for response definition
- Performance as a function of dR quality cut
- Check different truth jet matching schemes for response definition
- Evaluating the impact of soft lepton information (Prajita)
- Performance with LeptonID
- Apply additional selection on soft electron
- Auxiliary tasks (Brendon)
- Fixing weights of TruthOrigin classes
- Add TruthParticleID
- Model architecture (Brendon)
- Attention mechanisms
- Hyperparameter optimization
Samples and models on SDF
TDD output files:
# ttbar non-all hadronic sample in PHYSVAL format; 600 GB /gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples/nonallhad_PHYSVAL_mc20_410470 # ttbar samples in FTAG format; 900 GB /gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples/Rel22_ttbar_DiLep /gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples/Rel22_ttbar_SingleLep /gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples/Rel22_ttbar_AllHadronic
The following variables are available in the latest PHYSVAL TDD samples:
Umami-preprocessing datasets:
# UPP_1: ttbar non-all had FTAG1, 1.5mil jets; 1 GB /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/FTag_upp/Rel22_ttbar_AllHadronic_bjetonly # UPP_2: ttbar non-all had PHYSVAL, 100mil jets w/ 10 < pt_true; 133 GB /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/PHYSVAL_upp/Rel22_ttbar_NonAllHadronic/ # UPP_3: ttbar non-all had PHYSVAL, 49mil jets w/ 10 < pt_true < 50 GeV + 50mil jets w/ pt_true > 50 GeV; 112 GB /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/upp_output/physval/ttbar_allhad/half-half-100mil-bjet_resampled/
Trained models:
### Trained on UPP_1 # No auxiliary tasks /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-ftag1_1p5mil_NoAux_21param_20230918-T082841 /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-ftag1_1p5mil_NoAux_21param_20230918-T174839 ### Trained on UPP_2 # No auxiliary tasks, trained in 2 separate batches /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-physval_80mil_NoAux_21param_v1_20231013-T121459 /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-physval_80mil_NoAux_21param_v2_20231013-T121459 # With auxiliary tasks /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-physval_80mil_Aux_21param_20231023-T080935 ### Trained on UPP_3 /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/models/GNX-bjr-physval_80mil_NoAux_21param_PtResampled
SDF preliminaries
Migration to S3DF
In Jan/Feb 2024, we migrated from SDF to S3DF. Here are some helpful documentations:
Note that our installation of conda is at /sdf/group/atlas/sw/conda.
Compute resources
SDF has a shared queue for submitting jobs via slurm, but this partition has extremely low priority. Instead, use the usatlas partition or request Michael Kagan to join the atlas partition.
You can use the command sacctmgr list qos name=atlas,usatlas format=GrpTRES%125 to check the resources available for each:
atlas has 2 CPU nodes (2x 128 cores) and 1 GPU node (4x A100); 3 TB memoryusatlas has 4 CPU nodes (4x 128 cores) and 1 GPU node (4x A100, 5x GTX 1080. Ti, 20x RTX 2080 Ti); 1.5 TB memory
Environments
For sdf
An environment needs to be created to ensure all packages are available. We have explored some options for doing this.
Option 1: stealing instance built for SSI 2023. This installs most useful packages but uses python 3.6, which leads to issues with h5py.
Starting Jupyter sessions via SDF web interface
- SDF web interface > My Interactive Sessions > Services > Jupyter (starts a server via SLURM)
- Jupyter Instance > slac-ml/SSAI
Option 2 (preferred): create your own conda environment. Follow the SDF docs to use ATLAS group installation of conda.
There is also a slight hiccup with permissions in the folder /sdf/group/atlas/sw/conda/pkgs, which one can sidestep by specifying their own folder for saving packages (in GPFS data space).
The TLDR is:
# Add the following lines to ~/.condarc file (create default file with conda config) pkgs_dirs: - /gpfs/slac/atlas/fs1/d/<user>/conda_envs envs_dirs: - /gpfs/slac/atlas/fs1/d/<user>/conda_envs channels: - defaults - conda-forge
This will allow you to also create environments saved in GPFS rather than the login node, which fills up quickly, using –prefix /gpfs/slac/atlas/fs1/d/<user>/conda_envs/env_name .
conda init # the previous will be added to your bashrc file conda env create -f bjr_v01.yaml # for example(bjr_v01) conda install jupyter
This env can be activated when starting a kernel in Jupyter by adding the following under Custom Conda Environment:
export CONDA_PREFIX=/sdf/group/atlas/sw/conda
export PATH=${CONDA_PREFIX}/bin/:$PATH
source ${CONDA_PREFIX}/etc/profile.d/conda.sh
conda env list
conda activate bjr_v01
For the following packages (UPP, SALT, and puma) there are predefined conda environments available (made by Brendon). You can activate them with the usual conda activate command. They are here:
/gpfs/slac/atlas/fs1/d/bbullard/conda_env_yaml/ ├── puma.yaml ├── salt.yaml └── upp.yaml
Add the following to your .bashrc script as well
export PATH="/sdf/group/atlas/sw/conda/bin:$PATH"
export TMPDIR="${SCRATCH}"
For S3DF
Setting up the conda environment has steps similar to those of SDF. To use the atlas group conda, use following steps
1.) Add the following in your .bashrc
export PATH="/sdf/group/atlas/sw/conda/bin:$PATH"
2.) Add the following to your .condarc
pkgs_dirs: - /fs/ddn/sdf/group/atlas/d/<user>/conda_envs/pkgs envs_dirs: - /fs/ddn/sdf/group/atlas/d/<user>/conda_envs/envs channels: - conda-forge - defaults - anaconda - pytorch
3.) source conda.sh
> source /sdf/group/atlas/sw/conda/etc/profile.d/conda.sh
4.) cross check if conda is set correctly
> which conda
#following should appear if things went smoothly
conda ()
{
\local cmd="${1-__missing__}";
case "$cmd" in
activate | deactivate)
__conda_activate "$@"
;;
install | update | upgrade | remove | uninstall)
__conda_exe "$@" || \return;
__conda_reactivate
;;
*)
__conda_exe "$@"
;;
esac
}
5.) Now follow the same steps as SDF to create and activate the conda environment
Producing H5 samples with Training Dataset Dumper
Introduction
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Overview (FTag Workshop Sept. 2023): presentation by Nikita Pond
Tutorial: TDD on FTag docs
General introduction to Athena in Run 3 (component accumulator): ATLAS Software Docs
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
TDD productions:
v2.1: TDD code, BigPanda
Run with grid-submit -c configs/PHYSVAL_EMPFlow_jer.json -e AntiKt4EMPFlow_PtReco_Correction_MV2c10_DL1r_05032020.root -i BTagTrainingPreprocessing/grid/inputs/physval-jer.txt bjer
NOTE: you will need to copy the PtReco histogram into the top level of training-dataset-dumper
- Issue with datasets not being available on the grid.. need to request a new production
- GSC pre-recommendation (gitlab), jet calibrations in Athena
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
We are using a custom fork of training-dataset-dumper, which is used to extract h5 files from derivations - we are currently using FTAG1 / PHYSVAL / JETM2.
The fork in slac-bjr has our working code to dump datasets based on the AntiKt4TruthDressedWZJets container and EMPFlow as well as UFO jets.
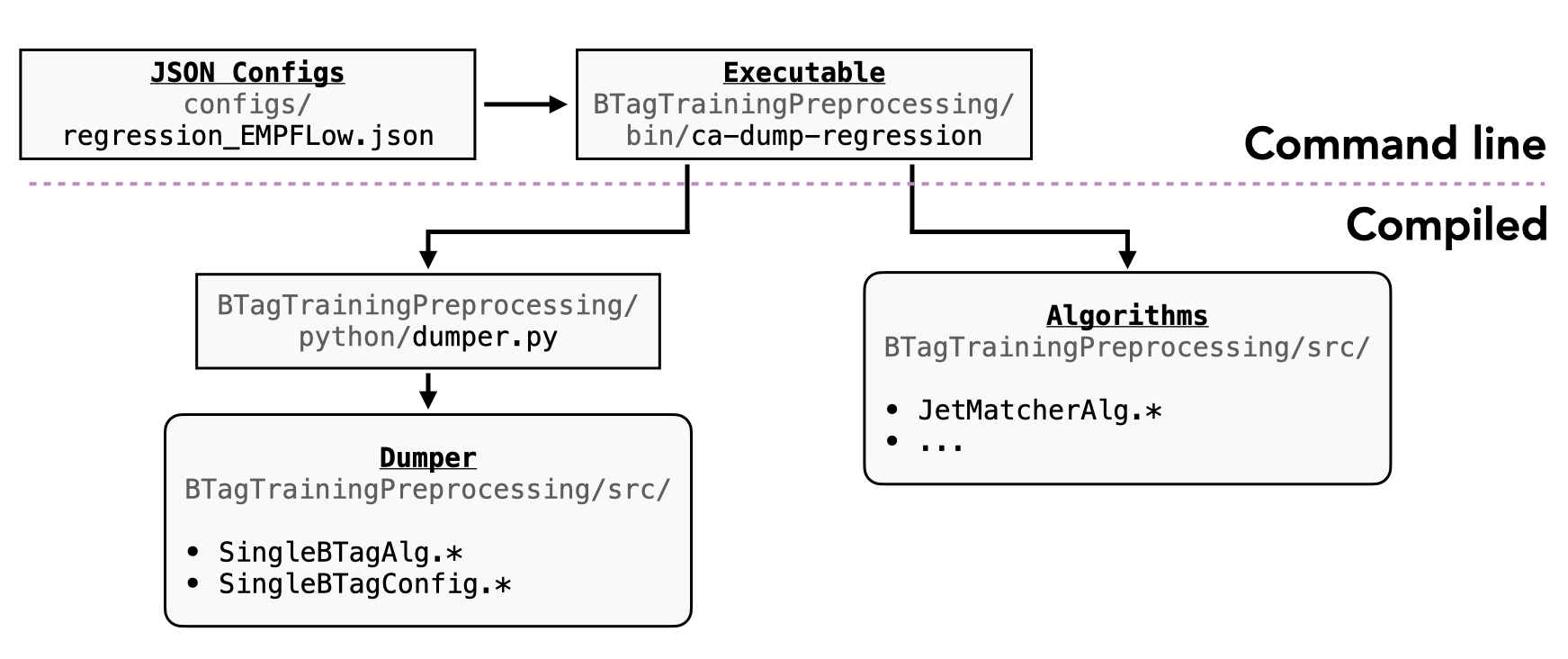
The main pieces of code that we are developing for the dumper are:
| Function | AntiKt4EMPFlowJets | AntiKt4UFOCSSKJets |
|---|---|---|
| Executable/Athena interface | BTagTrainingProcessing/bin/ca-dump-regression | |
| Top level configuration | configs/regression_EMPFlow*.json | configs/regression_UFO.json |
Class definitions:
- SingleBTagAlg.*: C++ entry point, called by dumper.py
- processSingleBTagEvent.*: templated method to apply tools based on config file
- SingleBTagTools:
- SingleBTagConfig:
We are currently using Athena release 23.0.26, this comes before Sam Van Stroud made a series of changes to the SoftElectronTruthDecoratorAlg in FTag tools that mostly broke things (in release 23.0.34 and 23.0.39). Note that Athena needs to be checked out, rather than AnalysisBase.
Component accumulator
The component accumulator currently uses two algorithms for b-jet regression:
- JetMatcherAlg: truth-reco jet matching
The following aliases can be used for convenient local testing:
alias test_PHYSVAL="ca-dump-regression -c configs/regression_EMPFlow.json -j AntiKt4EMPFlow --soft-lepton -o PHYSVAL.EMPflow.test.100.h5 -m 100 /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/PHYSVAL/mc20_13TeV/DAOD_PHYSVAL.29398576._003055.pool.root.1" alias test_JETM2_EMPFlow="ca-dump-regression -c configs/regression_EMPFlow.json -j AntiKt4EMPFlow -o JETM2.EMPflow.test.100.h5 -m 100 /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/JETM2/mc20_13TeV/DAOD_JETM2.35974512._003603.pool.root.1" alias JETM2_test_file="/gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/JETM2/mc20_13TeV/DAOD_JETM2.35974512._003603.pool.root.1" alias PHYSVAL_test_file="/gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/PHYSVAL/mc20_13TeV/DAOD_PHYSVAL.29398576._003055.pool.root.1"
Some predefined decorations are in TDD docs.
Local files
A set of test derivation files can be found at:
# FTAG1 mc21 (13.6 TeV) /gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/InputDAOD/mc21_13p6TeV.601230.PhPy8EG_A14_ttbar_hdamp258p75_dil.deriv.DAOD_FTAG1.e8453_s3873_r14372_p5627/DAOD_FTAG1.33108625._000137.pool.root.1 # PHYSVAL mc20 (13 TeV) /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/JETM2/mc20_13TeV/DAOD_JETM2.35974512._003603.pool.root.1 # JETM2 mc20 (13 TeV) [First test sample of new definition] /gpfs/slac/atlas/fs1/d/bbullard/BjetRegression/DAOD/PHYSVAL/mc20_13TeV/DAOD_PHYSVAL.29398576._003055.pool.root.1
The current set of available Ntuples is available on:
/gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples ├── nonallhad_PHYSVAL_mc20_410470 # High stats PHYSVAL derivation ├── Rel22_ttbar_AllHadronic ├── Rel22_ttbar_DiLep └── Rel22_ttbar_SingleLep
Adding Lepton & soft lepton to the TDD output
I (Prajita) am using the tools that already exist in the TDD.
Adding Electron and soft Electron
As shown below, two new algorithms are added to the same component accumulator. The first algo adds 10 electrons present near a jet (dR<0.4). The second and third lines add the algorithm that does soft electron tagging. The soft electron is defined as a single electron near a jet with the maximum probability of coming from a heavy flavor decay. This probability is evaluated by a e/gamma DNN tool.
sub.addEventAlgo(CompFactory.SingleBTagElectronAlg('electronAugmenter'))
sub.addEventAlgo(CompFactory.FlavorTagDiscriminants.SoftElectronDecoratorAlg('SoftElectronDecoratorAlg'))
sub.addEventAlgo(CompFactory.FlavorTagDiscriminants.SoftElectronTruthDecoratorAlg('SoftElectronTruthDecoratorAlg'))
Adding soft muon
Soft muon information is available as Aux variables in FTAG DAOD, and these can be added just by adding them to configs as "float" to "btag" variables.
example config of adding SoftElectron and Electron information. softmuon info also added in the same way as soft el.
Calibration/Selection to different objects
jets: config to calibration/selection
-Reco Jets
- rel 22 pre-recommendations
- baseline kinematic selection pT>20GeV & |eta|<2.5
- min_jvt 0.5
- event level jet cleaning
-truth jet selection
"pt_minimum": 2e3, "abs_eta_maximum": 2.5, "dr_maximum": 0.4
- 20 GeV truth jetpT applied at the umami preprocessing step
electrons:
Details of electron selection
for soft electron selection, "Loose" identification working point, and all of these selections.
soft muon:
Soft muon tagging is done in Athena by the FTAGSoftMuon Tagger tool. combined muon with basic kinematic selection pT > 5 GeV ‣ |η| < 2.5 ‣ d0 < 4 mm
Samples with electron, soft electron, and soft muon information
base_sdf_dir/nonallhad_PHYSVAL_mc20_410470 This was processed on mc20 all hadronic ttbar samples, to be replaced by mc21 or mc23 13p6 samples.
Plotting with Umami/Puma (Work In Progress)
NOTE: we have not yet used umami/puma for plotting or preprocessing, but it can in principle be done and we will probably try to get this going soon.
Plotting with umami
Umami (which relies on puma internally) is capable of producing plots based on yaml configuration files.
The best (read: only) way to use umami out of the box is via a docker container. To configure on SDF following the docs, add the following to your .bashrc:
export SDIR=/scratch/${USER}/.singularity
mkdir $SDIR -p
export SINGULARITY_LOCALCACHEDIR=$SDIR
export SINGULARITY_CACHEDIR=$SDIR
export SINGULARITY_TMPDIR=$SDIR
To install umami, clone the repository (which is forked in the slac-bjr project):git clone ssh://git@gitlab.cern.ch:7999/slac-bjr/umami.git
Now setup umami and run the singularity image:
source run_setup.sh singularity shell /cvmfs/unpacked.cern.ch/gitlab-registry.cern.ch/atlas-flavor-tagging-tools/algorithms/umami:0-19
Plotting exercises can be followed in the umami tutorial.
Plotting with puma standalone
Puma can be used to produce plots in a more manual way. To install, there was some difficulty following the nominal instructions in the docs. What I (Brendon) found to work was to do:
conda create -n puma python=3.11 pip install puma-hep pip install atlas-ftag-tools
This took quite some time to run, so (again) save yourself the effort and use the precompiled environments.
Pre-processing
SALT likes to take preprocesses data file formats from Umami (though in principle the format is the same as what's produced by the training dataset dumper).
An alternative (and supposedly better way to go) than using umami for preprocessing is to use the standalone umami-preprocessing (UPP).
Follow the installation instructions on git. This is unfortunately hosted on GitHub, so we cannot add it to the bjr-slac GitLab group A working fork
is available under Brendon's account and contains UPP configs for each sample configuration (bjr branch). These do not yet implement resampling, but there are 2 methods that can be explored (resampling and reweighting).
You can cut to the chase by doing conda activate upp, or follow the docs for straightforward installation.
To do the preprocessing to make the training, validation, and test sets, do:
preprocess --config configs/bjr.yaml --split all
Notes on logic of the preprocessing config files
There are several aspects of the preprocessing config that are not documented (yet). Most things are self explanatory, but note that:
- within the FTag software, there exist flavor classifications (for example lquarkjets, bjets, cjets, taujets, etc). These can be used to define different sample components. Further selections
based on kinematic cuts can be made through theregionkey. - It appears only 2 features can be specified for the resampling (nominally pt and eta).
- The binning also appears not to be respected for the variables used in resampling.
Model development with SALT
The slac-bjr git project contains a fork of SALT. One can follow the SALT documentation for general installation/usage. Some specific notes can be found below:
Creating conda environment
One can use the environment salt that has been setup in /gpfs/slac/atlas/fs1/d/bbullard/conda_envs,
otherwise you may need to use conda install -c conda-forge jsonnet h5utils. Note that this was built using the latest master on Aug 28, 2023.
SALT also uses comet for logging, which has been installed into the salt environment (conda install -c anaconda -c conda-forge -c comet_ml comet_ml).
Follow additional instructions for configuration on the SALT documentation.
If using the latest main branch from Dec 11 2023, after SALT model refactoring has occurred, there are some issues with CUDA version compatibility with SDF.
PyTorch v2.0.1 used in SALT requires newer CUDA version than is available (11.4). One may need to do the following:
conda uninstall pytorch conda install pytorch torchvision torchaudio cudatoolkit=11.4 -c pytorch
Note that the following error may appear if following the normal installation:
ImportError: cannot import name 'COMMON_SAFE_ASCII_CHARACTERS' from 'charset_normalizer.constant'
To solve it, you can do the following (again, this is already available in the common salt environment):
conda install chardet conda install --force-reinstall -c conda-forge charset-normalizer=3.2.0
Interactive testing
It is suggested not to use interactive nodes to do training, but instead to open a terminal on an SDF node by:
- Starting a Jupyter notebook (with a conda environment that has jupyter notebook installed, not necessarily the SALT conda env).
A singularity container used in the jupyter notebook will propagate to the terminal session, which is incompatible with SALT. - New > Terminal. From here, you can test your training configuration.
There are some additional points to be aware of:
- Running a terminal on an SDF node will reduce your priority when submitting jobs to slurm
- Be careful about the number of workers you select (in the PyTorch trainer object) which should be <= the number of CPU cores you're using (using more CPU cores parallelizes the data loading,
which can be the primary bottleneck in training) - The number of requested GPUs should match the number of devices used in the training.
- The number of jets you use in the test training should be larger then the batch size (2x works fine)
The following code can be used to test SALT training (it's basically the same thing you have in the slurm submission script):
salt fit -c configs/<my_config>.yaml --data.num_jets_train <small_number> --data.num_workers <num_workers>
For training on slurm
See the SALT on SDF documentation (also linked at the top of this page) and example configs in the SALT fork in the slac_bjr GitLab project.
For additional clarity, see the following description of the submission scripts. Change the submit_slurm.sh script as follows
#SBATCH --output="<your_output_path>/out/slurm-%j.%x.out" #SBATCH --error="<your_output_path>/out/slurm-%j.%x.err export COMET_API_KEY=<your_commet_key> export COMET_WORKSPACE=<your_commet_workspace> export COMET_PROJECT_NAME=<your_project_name> cd <your_path_to_salt_directory>
from top level salt directory can use following command to launch a slurm training job in sdf
sbatch salt/submit_slurm_bjr.sh
Both slurm-%j.%x.err andslurm-%j.%x.out files will start to fill up
You can use standard sbatch commands from SDF documentation to understand the state of your job.
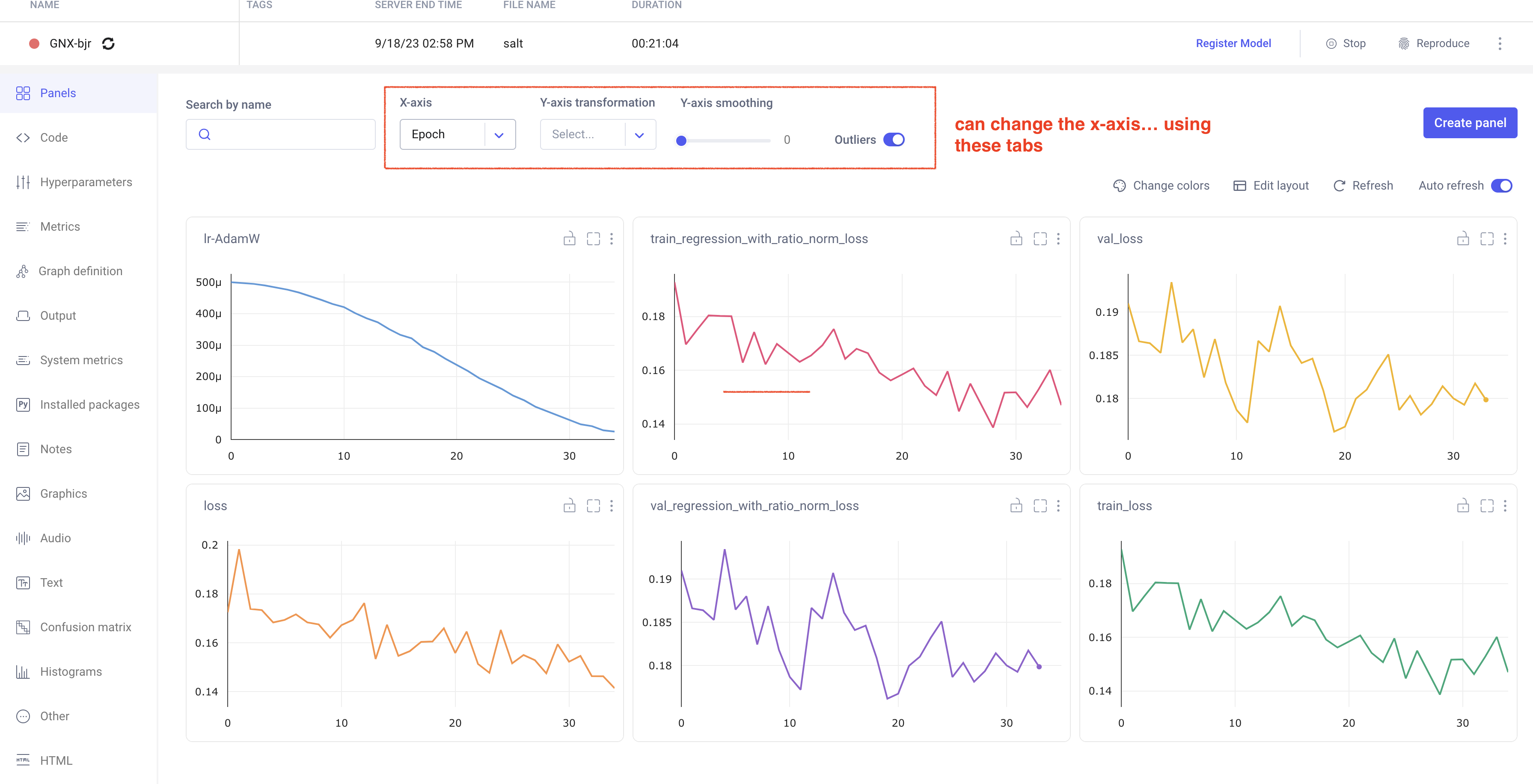
Comet Training Visualization
In your comet profile, you should start seeing the live update for the training which looks as follows. The project name you have specified in the submit script appears under your
workspace which you can click to get the graphs of live training updates.
Training Evaluation
Follow salt documentation to run the evaluation of the trained model in the test dataset. There is a separate batch submission script used for the model evaluation, but is very similar to what is used in the model training batch script.
The main difference is in the salt command that is run (see below). It will produce a log in the same directory as the other log files, and will produce a new output h5 file alongside the one you pass in for evaluation.
The important points are the following:
- --data.test_suff: Specify a suffix for the sample that is produced from the PredictionWriter callback specified in the model config. There is a separate list of features to be saved to this new file, along with the model output,
that can be used for studies of the model performance - --data.num_workers: you should use the same number of workers for the evaluation as for the training, since both are bottlenecked by the loading of the data
- --data.test_file: technically can be either the training, testing, or evaluation h5 file. In principle the testing file is created for this purpose. Philosophically, you want to keep this dataset separate so that you don't induce some kind of bias as you manually determine the hyper parameter optimization
- --ckpt_path: the specific model checkpoint you want to use. Out of the box, SALT should be picking the checkpoint with the lowest validation loss, but this has been found to not be very reliable. So always do it manually to be sure you know what model state you are actually studying.
srun salt test -c logs/<model>/config.yaml --data.test_file <path_to_file_for_evaluation>.h5 --data.num_jets_train <num_jets> --data.num_workers 20 --trainer.devices 1 --data.test_suff eval --ckpt_path logs/<model>/ckpts/epoch=<epoch>.ckpt
We are developing some baseline evaluation classes in the analysis repository to systematically evaluate model performance.
To do: develop this, and add some notes on it (Brendon/Prajita)
Miscellaneous tips
You can grant read/write access for GPFS data folder directories to ATLAS group members via the following (note that this does not work for SDF home folder)
groups <username> # To check user groups cd <your_directory> find . -type d|xargs chmod g+rx # Need to make all subdirectories readable *and* executable to the group
You can setup passwordless ssh connection following this page to create an ssh key pair. Then add this block to your ~/.ssh/config file (allows to tunnel directly to iana, but you can adapt to other hosts):
Host sdfiana HostName iana User <yourusername> IdentityFile ~/.ssh/<your private key> proxycommand ssh s3dflogin.slac.stanford.edu nc %h %p ForwardX11 yes Host s3dflogin.slac.stanford.edu
Discussion Summary
Suggestions from M. Kagan Jan 23, 2023
- Not to regress the neutrino pT directly, as the pT distribution might not be unimodal. Instead, try two different approaches
- Quantile regression with the median
- Auxilary task based on classification of neutrino pT class prediction, i.e., predict the bin 0-1 for the ratio of neutrino pT & jet pT. Also, add overflow bin
- loss function based on median rather than mean square error, ie, MAE rather than MSE