ami-local -b 1 -f interval=1 psana://exp=tmoc00118,run=222,dir=/cds/data/psdm/prj/public01/xtc |
Mask (2D or 3D array) is created mainly from calibration constants of particular detector.
For example, bring control nodes to the flowchart and connect their terminals as shown below.
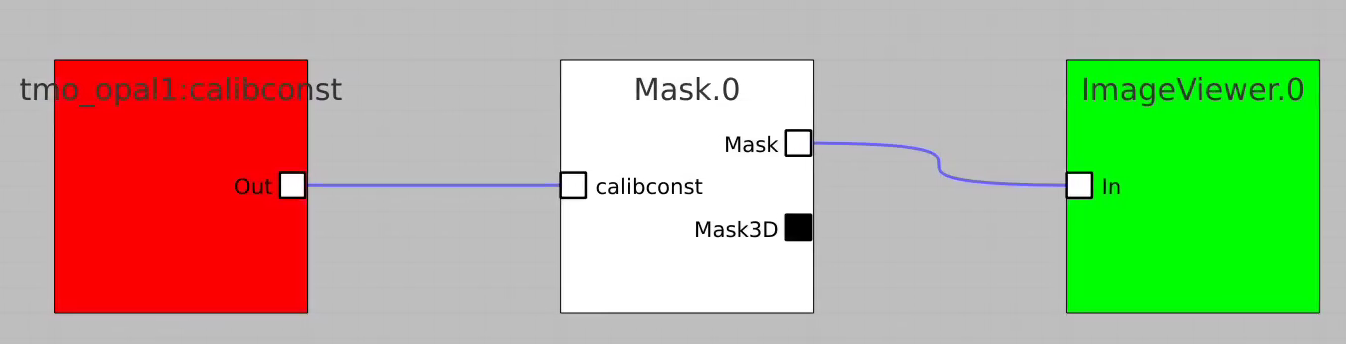
Click on "Apply" button, then click on Mask.0 and ImageViewer.0 control nodes to open editor for mask parameters and mask image viewer.
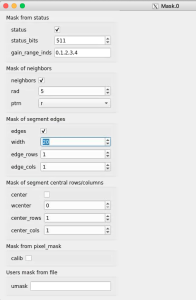
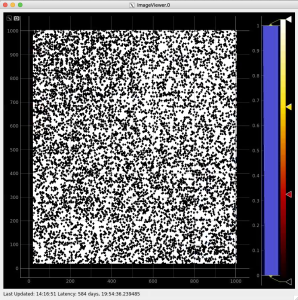
Adjust the mask parameters using editor window and click on "Apply" button again. Mask image will have changed according to set parameters.
ami-local -b 1 -f interval=1 psana://exp=uedcom103,run=7,dir=/cds/data/psdm/prj/public01/xtc |
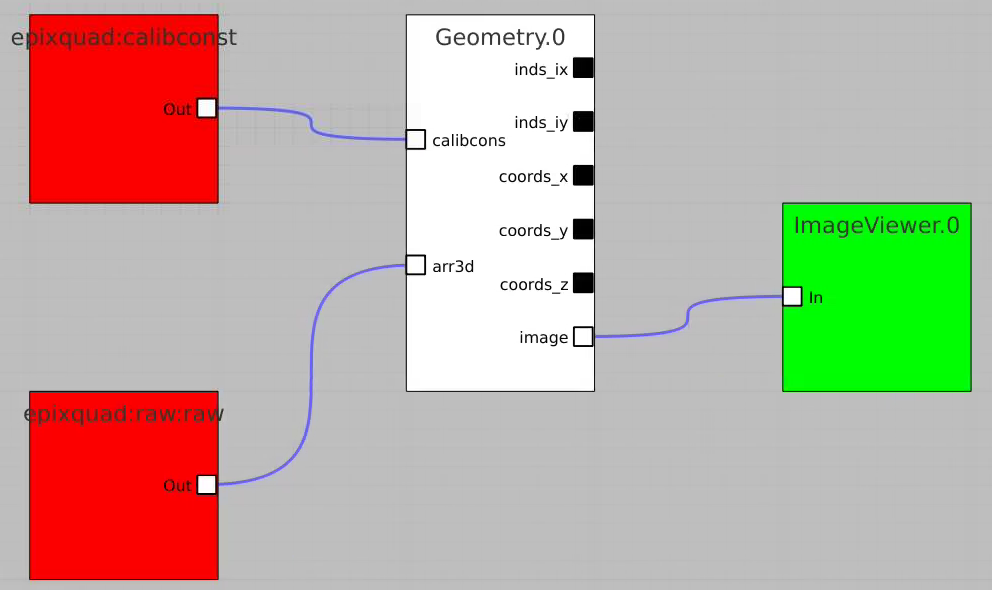
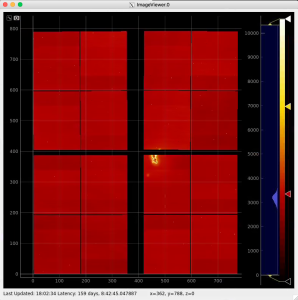
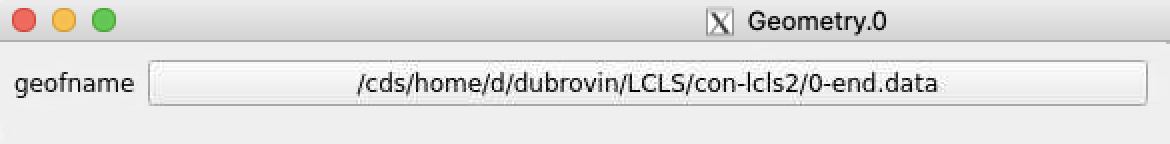
By default geometry data comes from DB associated with experiment or detector. Optional geometry file can be specified through the click on Geometry Control Node to deal with parameters window.
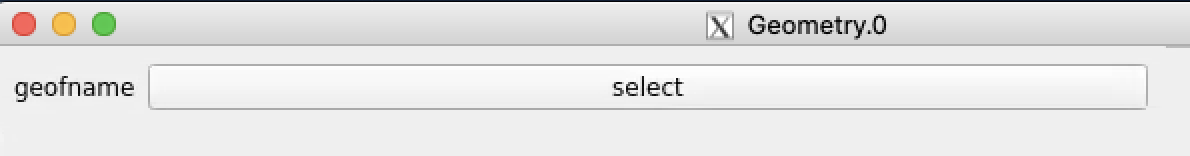
Click on "select" button and select desired geometry file, e.g.
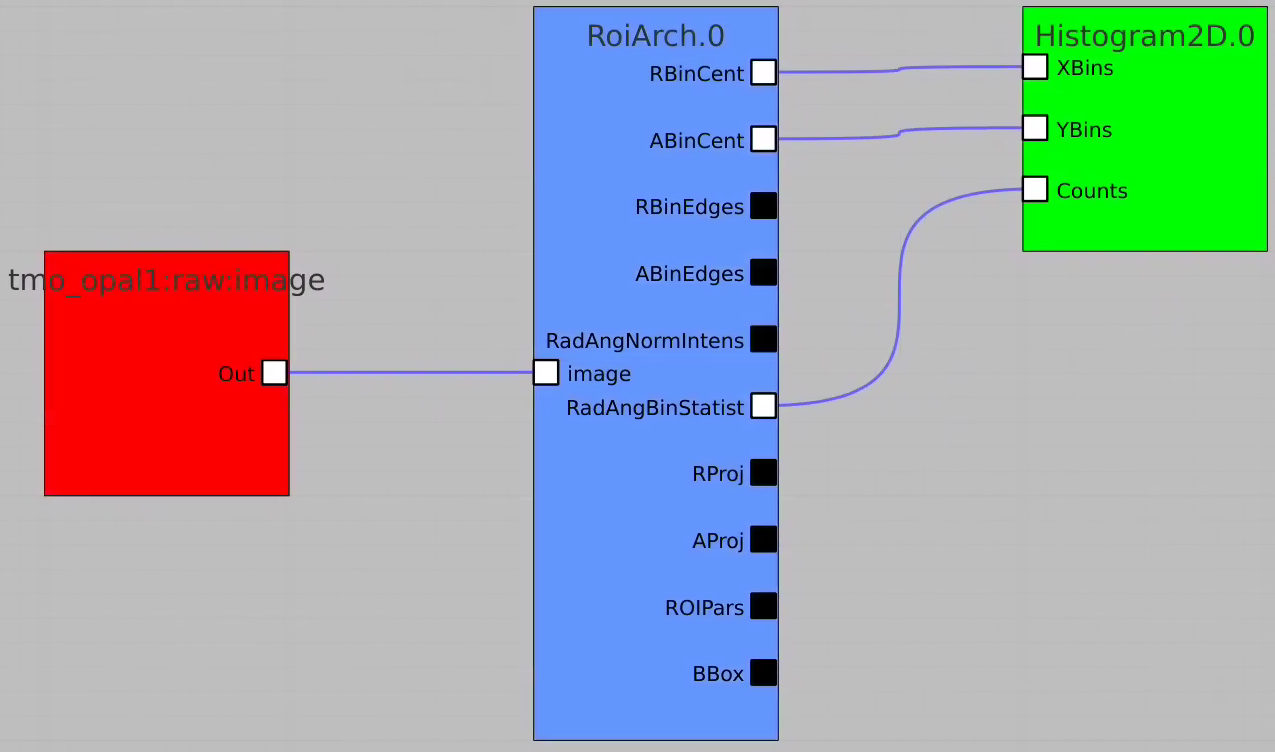
Add the Geometry Control Node as usually and remove input terminal for arr3d. Click on "Apply" and use any geometry output parameters except "image".

In simple case we use 2D image as input array without mask and ImageViewer with bin numbers in stead of actual scales for radial and angular dimensions.
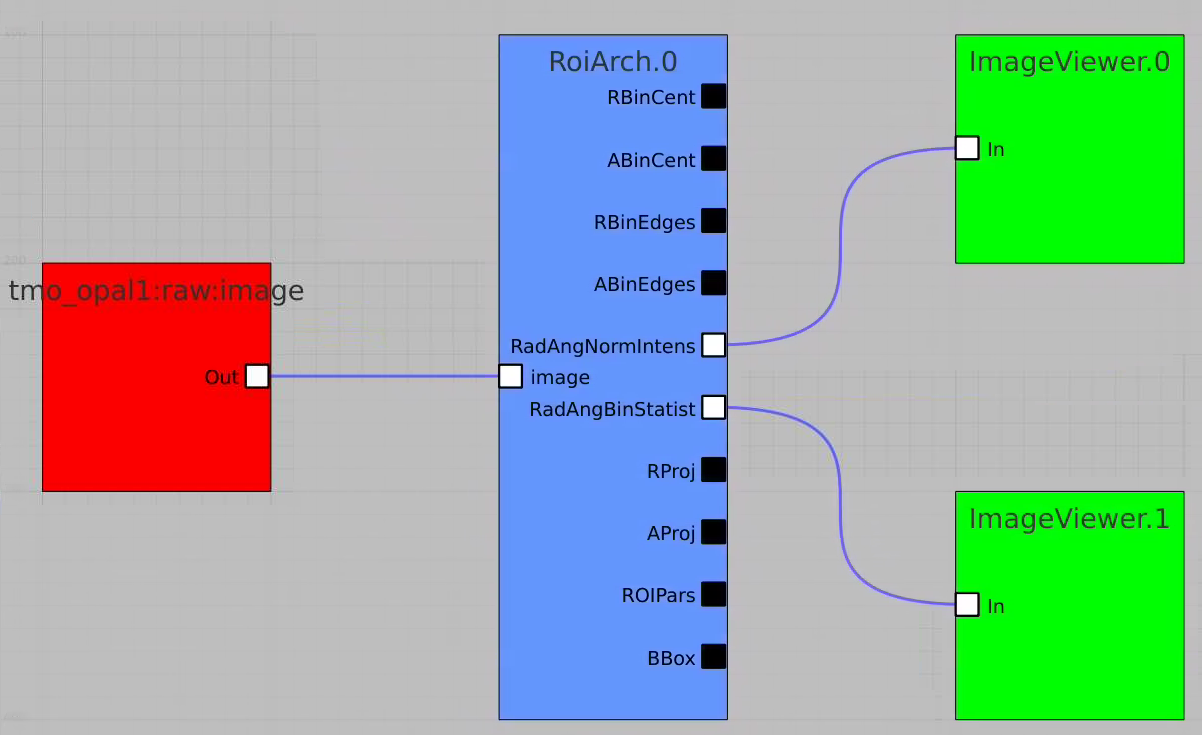
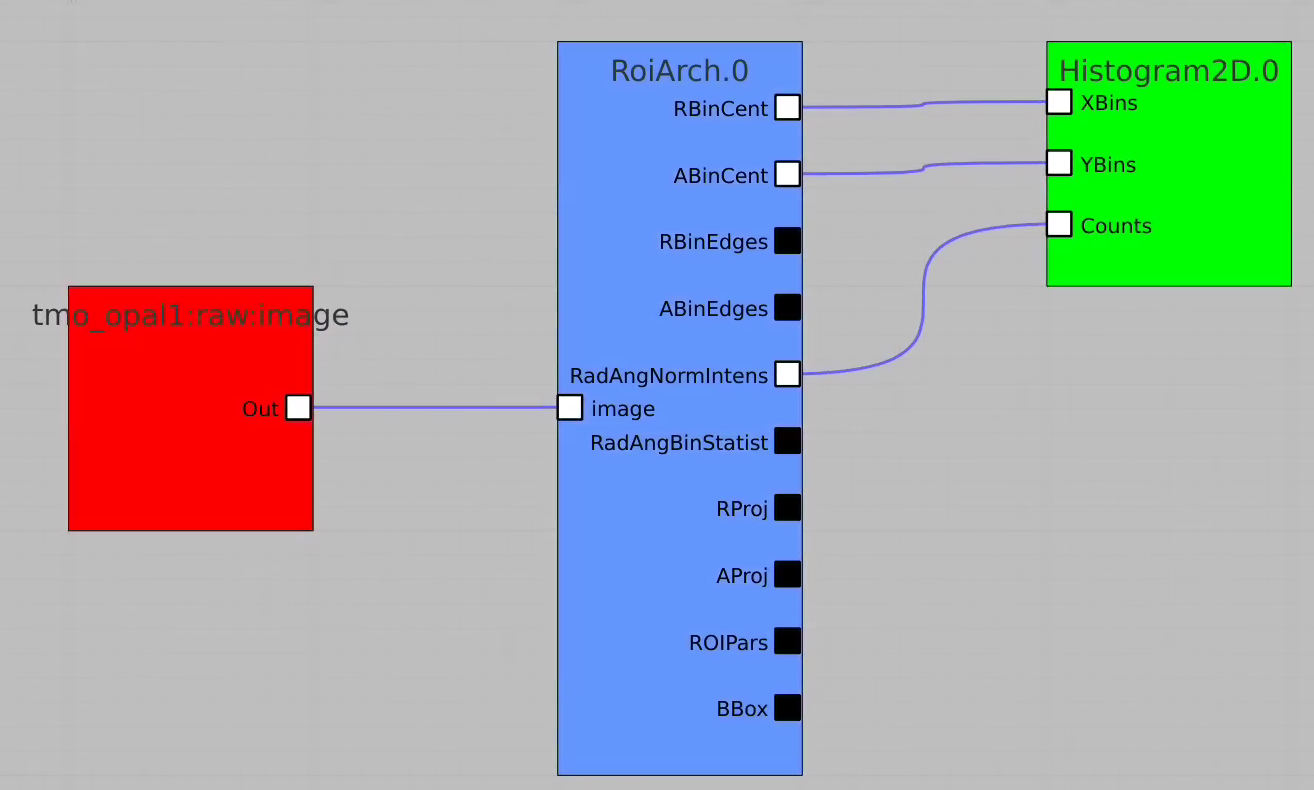
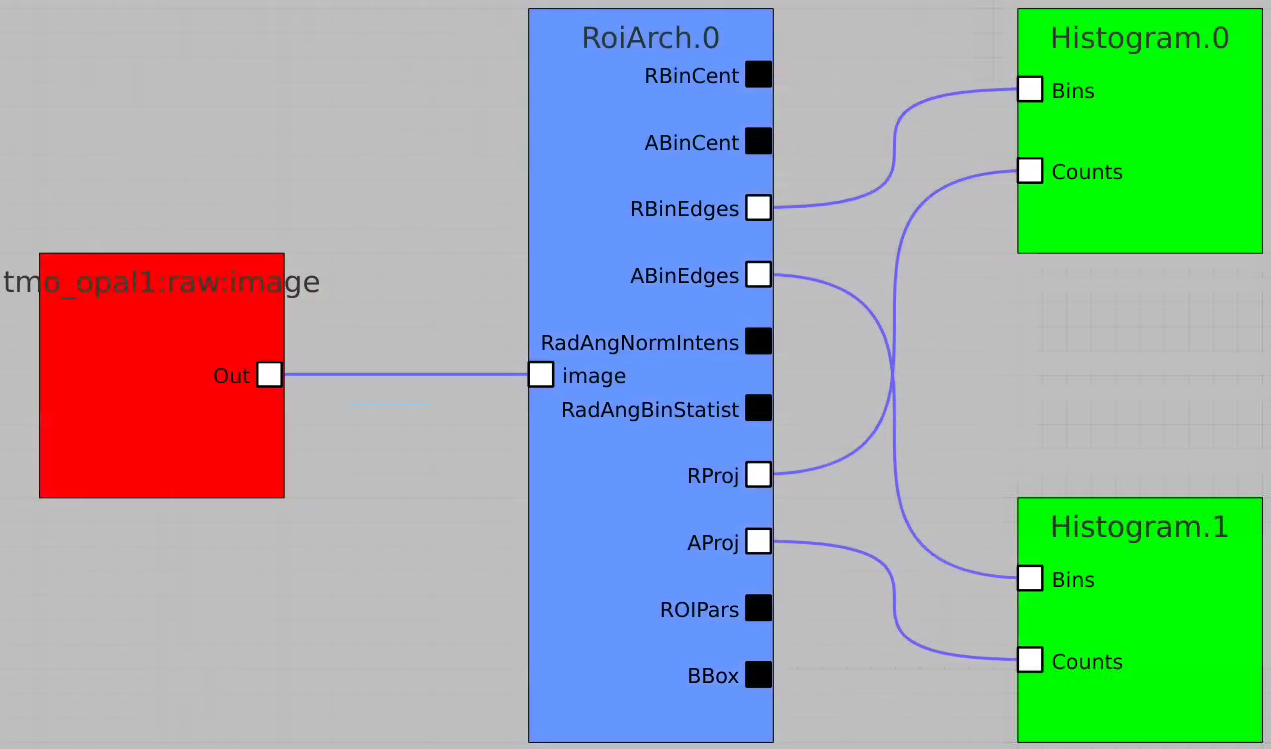
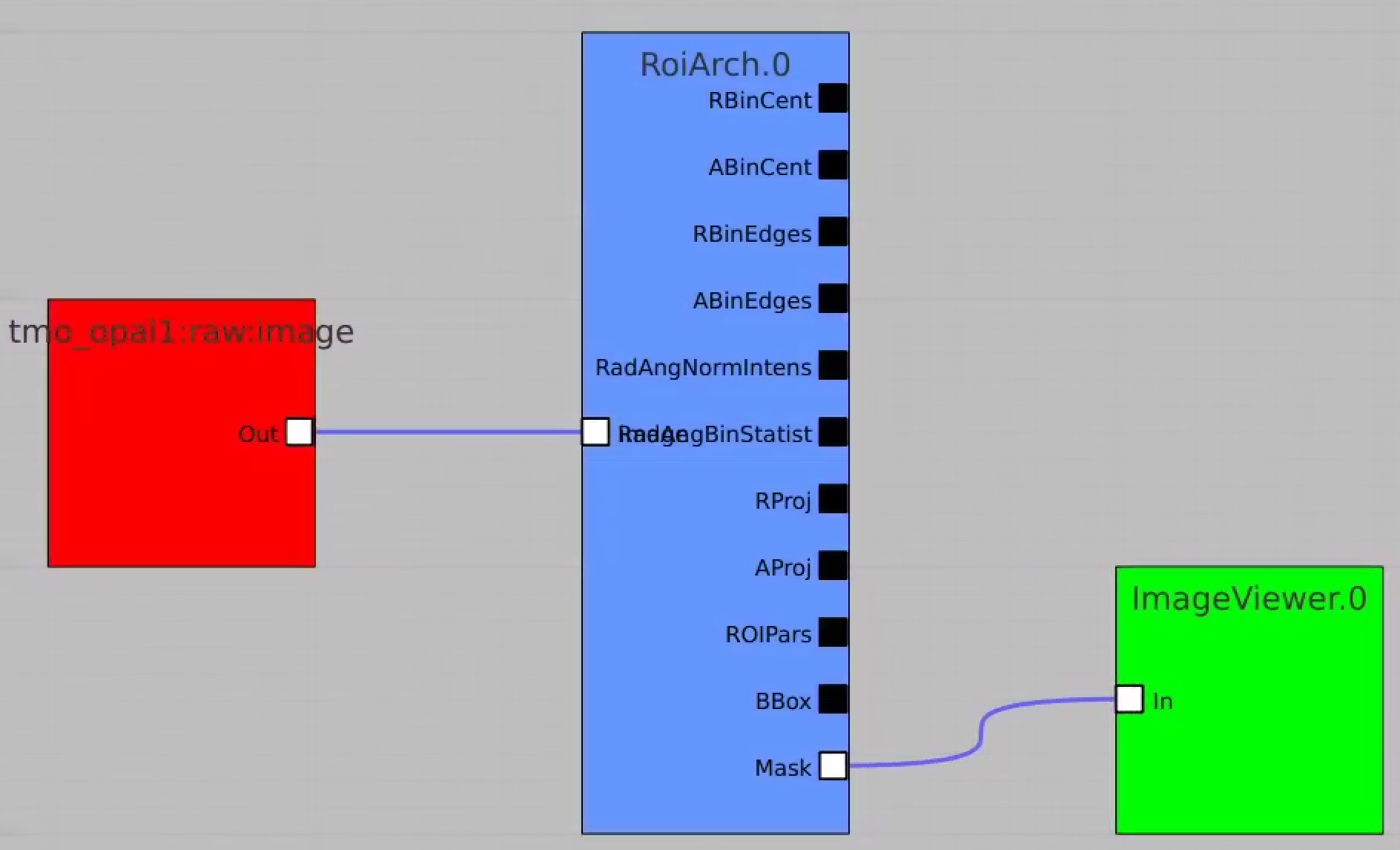
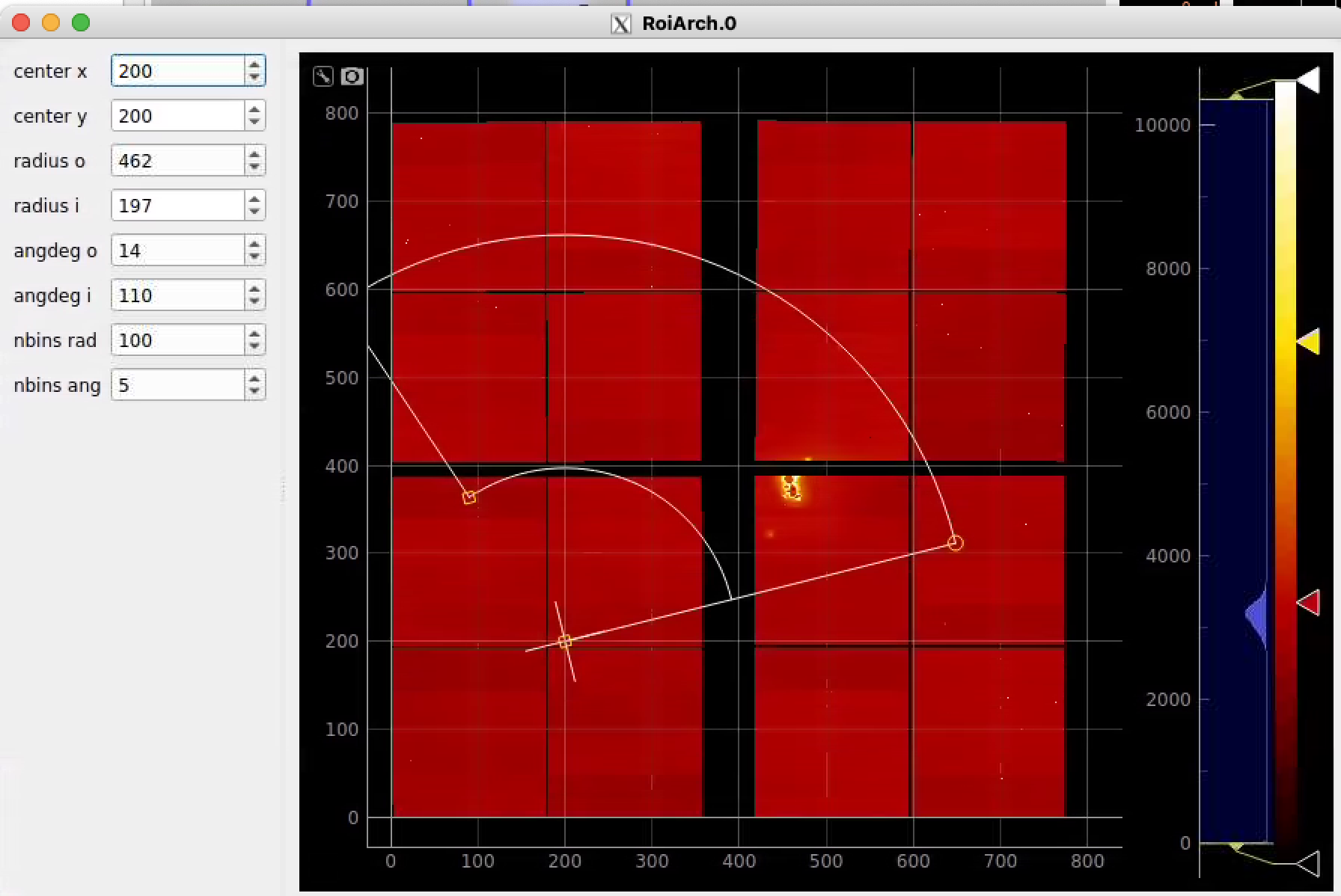
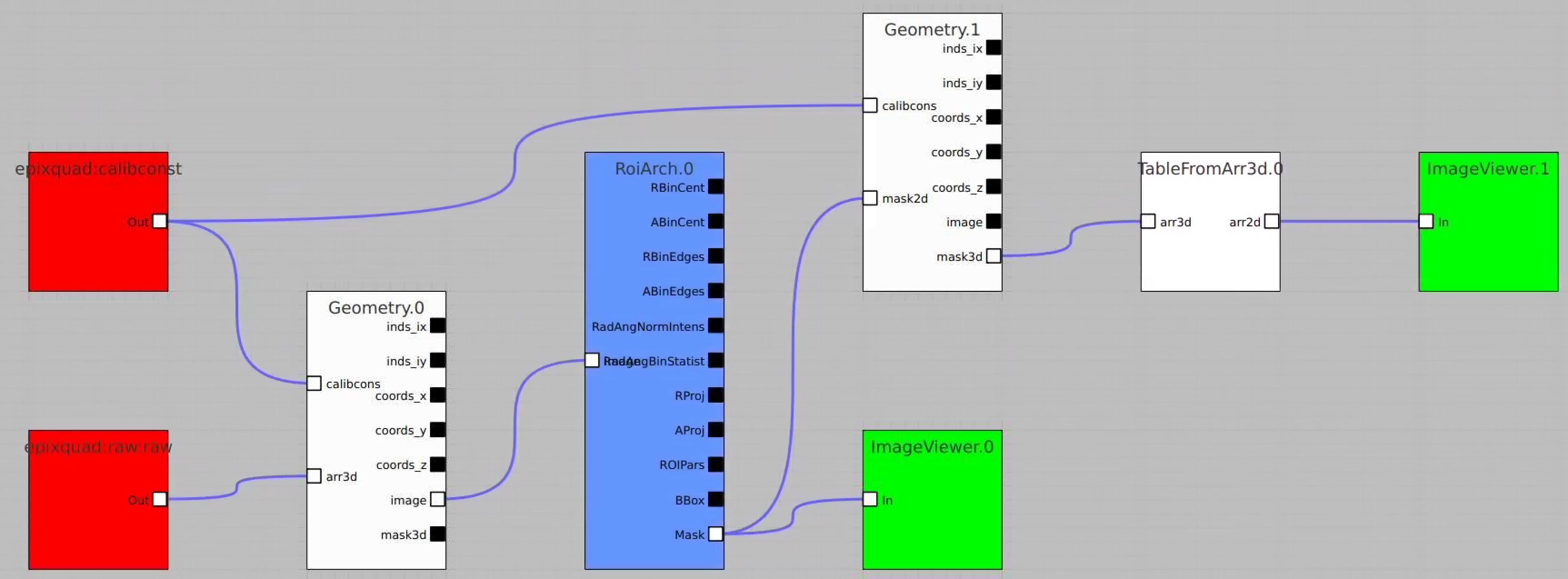
First, bring RoiArch to the flowchart, right-click on input "mask" terminal and select "Remove terminal" on pop-up window. (Currently ami2 does not work with optional terminals).

Bring other control nodes to the flowchart and connect them as shown below and click on "Apply" button.
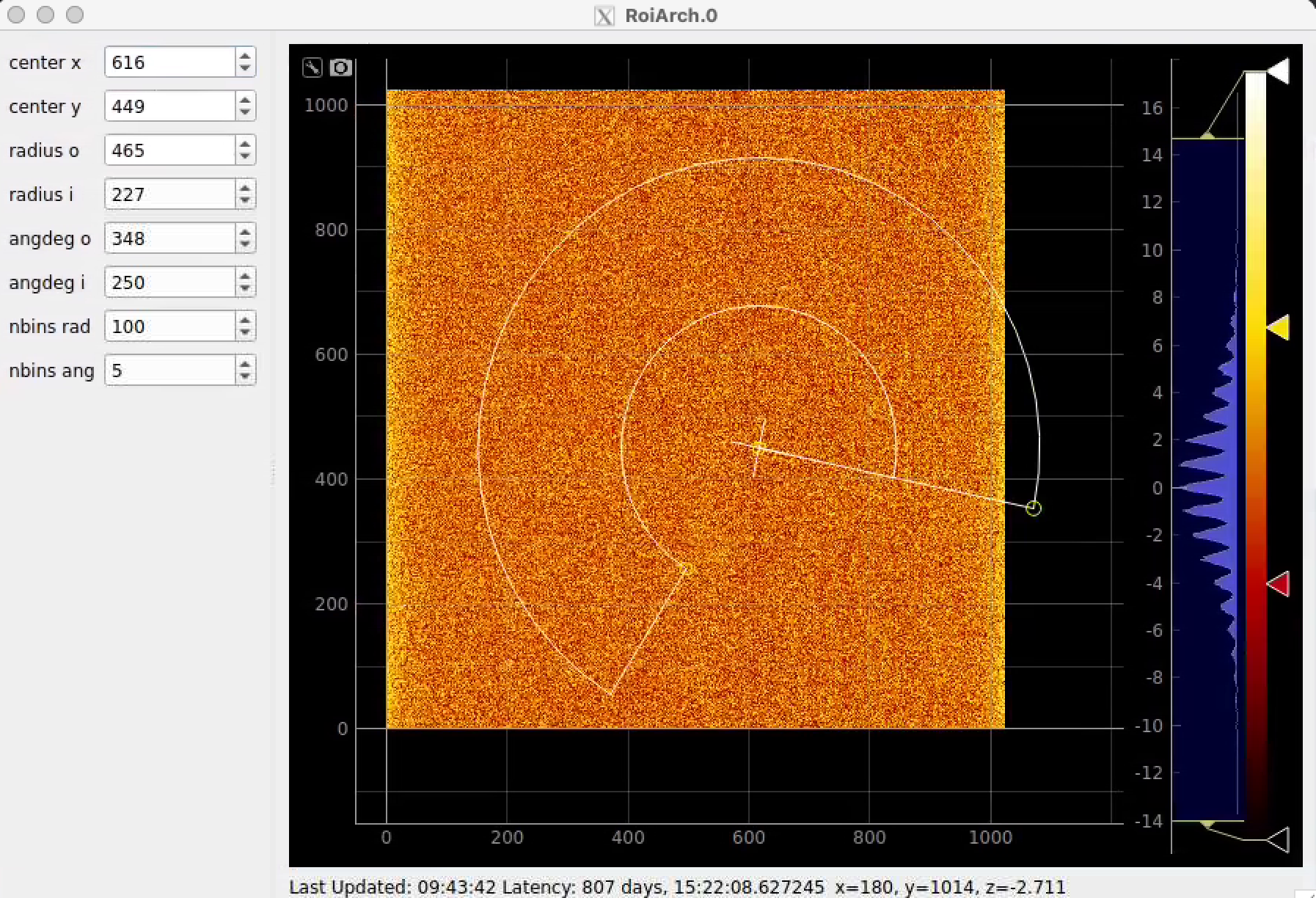
Click on RoiArch control node and adjust desired ROI region on pop-up window.
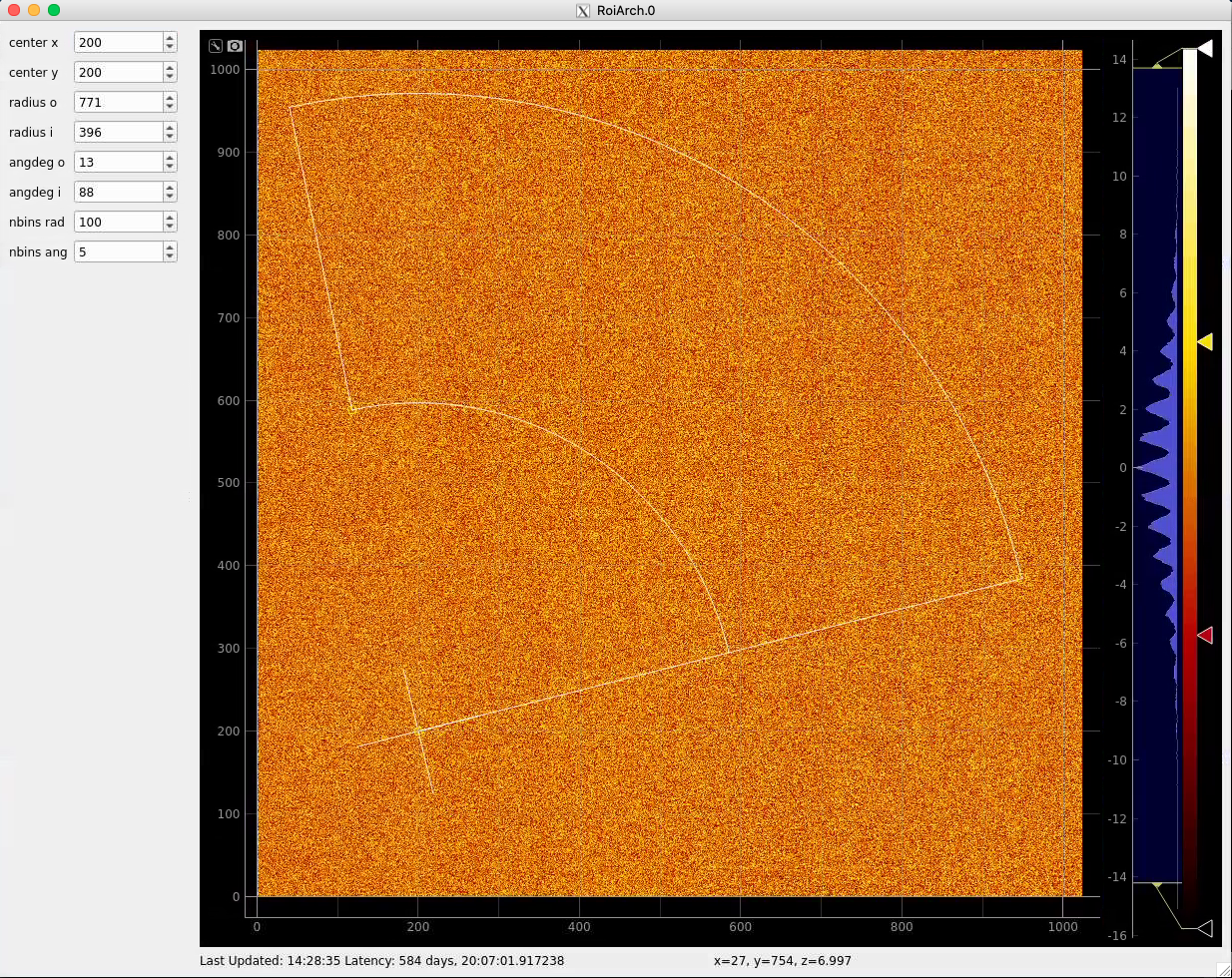
Image can be moved and zoomed by click-and-drag and scrolling mouse, respectively.
ArchROI can be adjusted using 3 control points for center, external radius and entire rotation, internal radius and arch angular size. Alternatively, ArchRoi parameters can be adjusted using editor for parameters in the left side of the window. Click on "Apply" button after mask parameters are set.
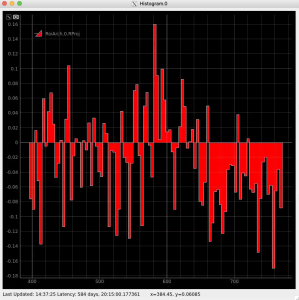
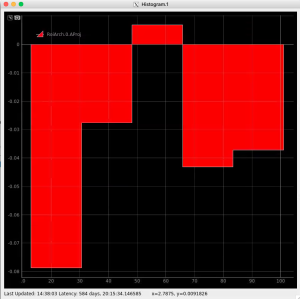
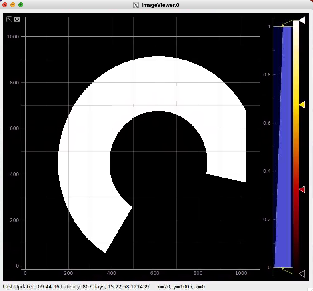
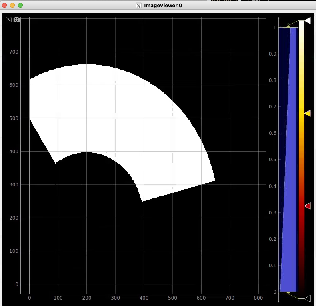
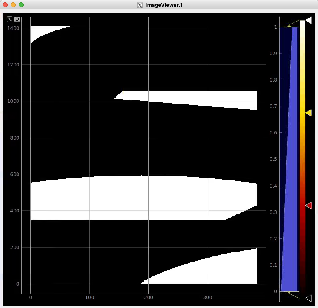
Then click on ImageViewer.0 and ImageViewer.1 control nodes to see images for r-angle per pixel normalized intensity and associated pixel per bin statistics.
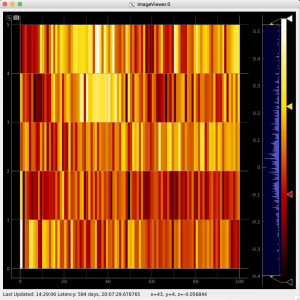
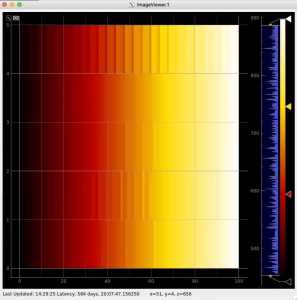
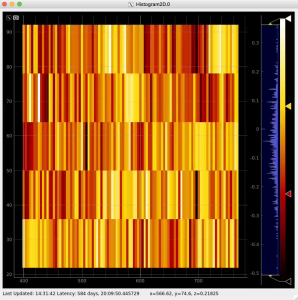
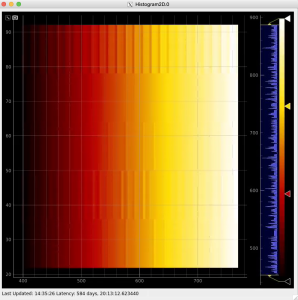
Arrays of N+1 bin edges should be used for 1D histograms
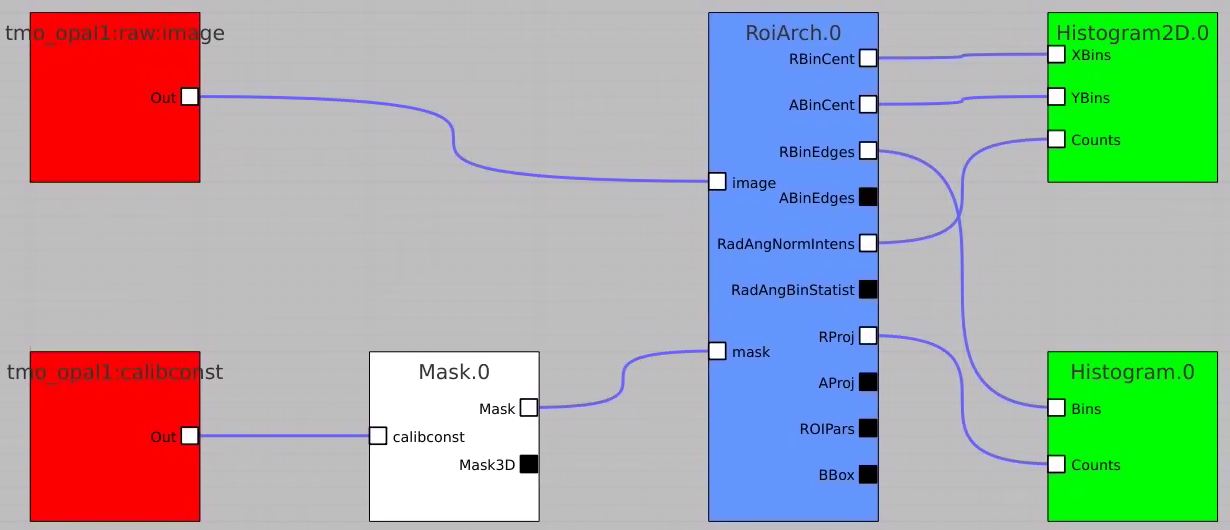
Bring RoiArch to the flowchart, connect th Mask.0 control node output (2D) Mask with RoiArch "mask" input terminal.
Then build other control nodes as in previous examples, click on "Apply" button, etc.
ami-local -b 1 -f interval=1 psana://exp=tmoc00118,run=222,dir=/cds/data/psdm/prj/public01/xtc |
ami-local -b 1 -f interval=1 psana://exp=uedcom103,run=7,dir=/cds/data/psdm/prj/public01/xtc |
NEW:
Example shows how to
AMI issues
Essential code is in ami/flowchart/library/Psalg.py ami-local -b 1 -f interval=1 psana://exp=uedcom103,run=7,dir=/cds/data/psdm/prj/public01/xtc |
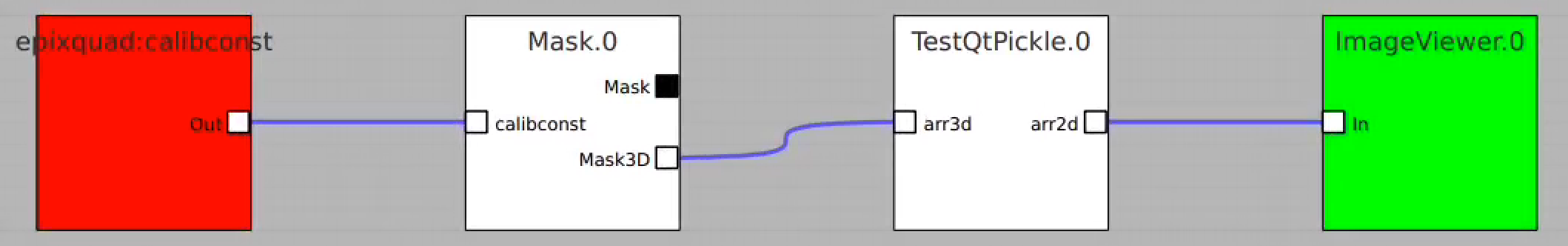
Crash:
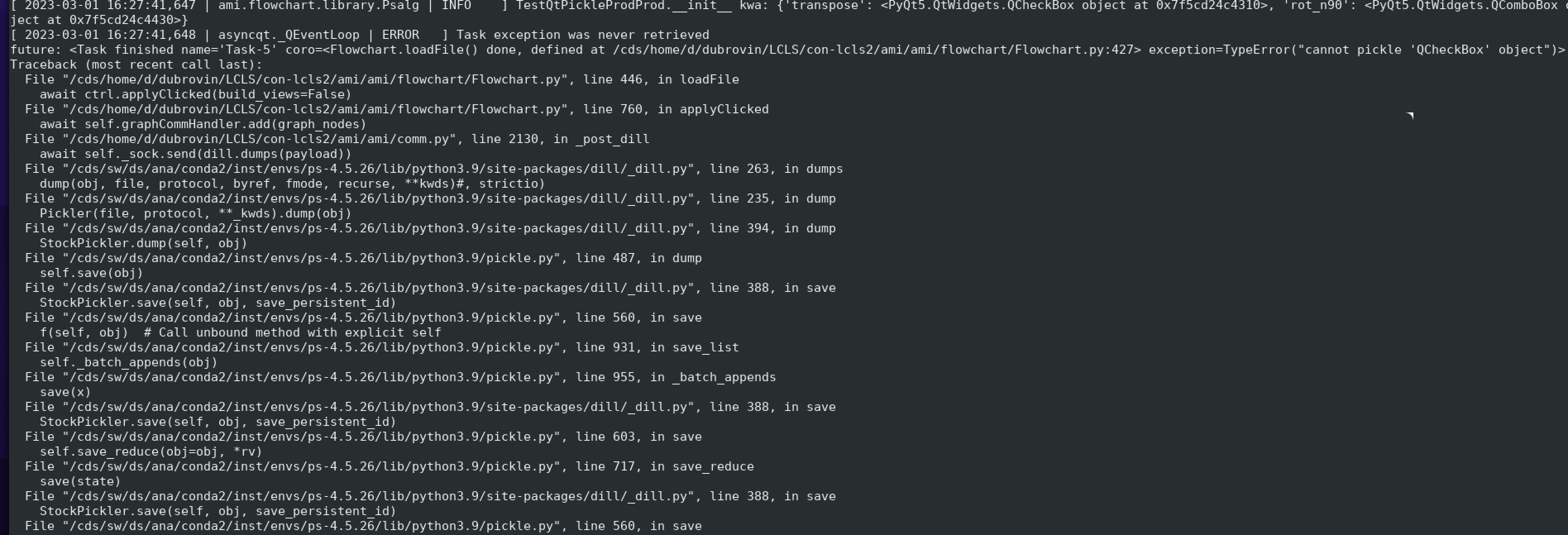
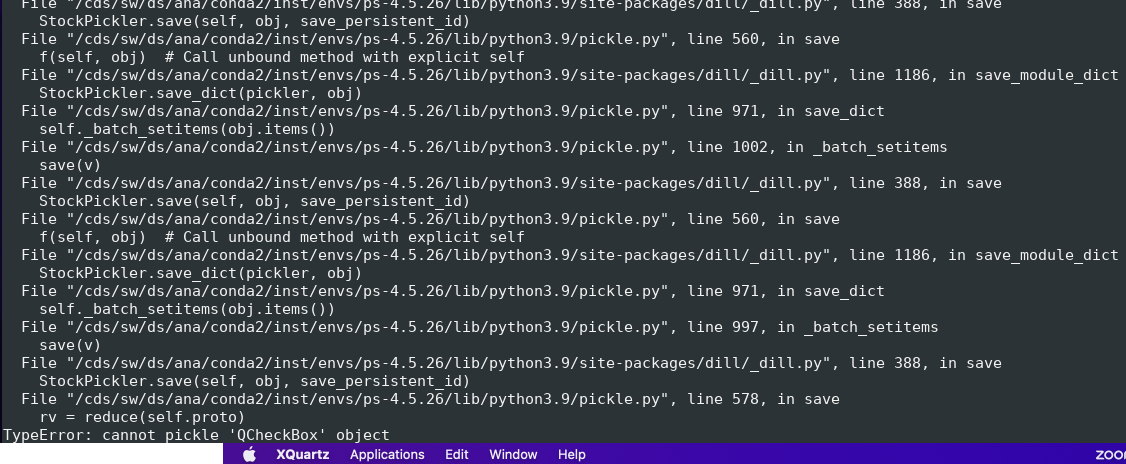
...
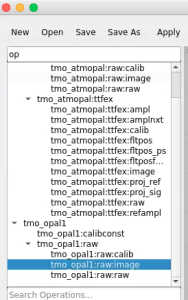
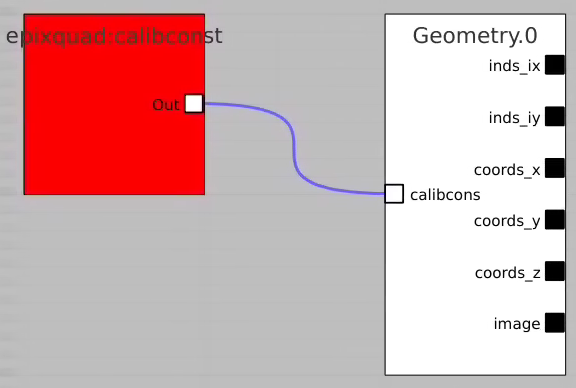
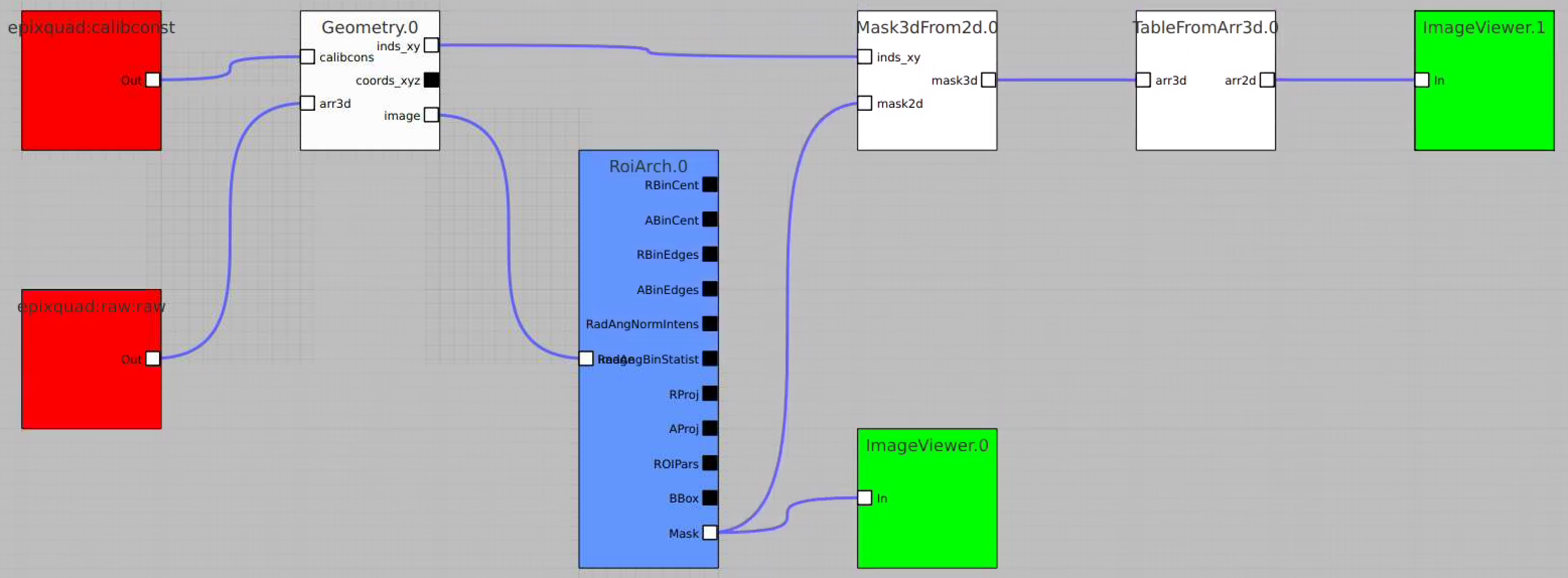
This example shows how to access in ami all components of the det.raw.calib(...) method such as calibration constants, per-panel trbit and asicPixelConfig, per-event gain factors and pedestals, mask, common mode correction, etc.
# for epixquad - 4-panel epix10ka: ami-local -b 1 -f interval=1 psana://exp=ueddaq02,run=569,dir=/cds/data/psdm/prj/public01/xtc # for single panel epixhr: ami-local -b 1 -f interval=1 psana://exp=rixx45619,run=121,dir=/cds/data/psdm/prj/public01/xtc |
In this example the PythonEditor box receives input objects for raw (np.ndarray), config, calibconst, accesses and prints internal components of the det.raw.calib(...) method and returns 2-d image for one retrieved arrays. For imaging, the 3-d array of common mode correction is converted to 2-d table of segments.
import psana.detector.utils_calib_components as ucc
np, info_ndarr, table_nxn_epix10ka_from_ndarr =\
ucc.np, ucc.info_ndarr, ucc.psu.table_nxn_epix10ka_from_ndarr
class EventProcessor():
def __init__(self):
self.counter = 0
self.cc = None
def begin_run(self):
pass
def end_run(self):
pass
def begin_step(self, step):
pass
def end_step(self, step):
pass
def on_event(self, raw, config, calibconst, *args, **kwargs):
self.counter += 1
if self.cc is None:
self.cc = ucc.calib_components_epix(calibconst, config)
cc = self.cc
kwa = {'status': True}
cmpars = (0, 7, 300, 10)
ctypes = cc.calib_types() # list of calibration types
npanels = cc.number_of_panels() # number of panels/segments in the detector
peds = cc.pedestals() # OR cc.calib_constants('pedestals')
gain = cc.gain() # OR cc.calib_constants('pixel_gain') # ADU/keV
gfactor = cc.gain_factor() # keV/ADU
status = cc.status() # 4-d array of pixel_status constants
comode = cc.common_mode() # tuple of common mode correction parameters
trbit_p0 = cc.trbit_for_panel(0) # list of per-ASIC trbit for panel 0
ascfg_p0 = cc.asicPixelConfig_for_panel(0) # matrix of asicPixelConfig for panel 0
mask = cc.mask(**kwa) # mask defined by kwa
dettype = cc.dettype() # detector type, e.g. "epix10ka" or "epixhr"
cbitscfg = cc.cbits_config_detector()
cbitstot = cc.cbits_config_and_data_detector(raw, cbitscfg)
gmaps = cc.gain_maps_epix(raw) # gain map
pedest = cc.event_pedestals(raw) # per-pixel array of pedestals in the event
factor = cc.event_gain_factor(raw) # per-pixel array of gain factors in the event
calib0 = cc.calib(raw, cmpars, **kwa) # method calib
cmcorr = cc.common_mode_correction(raw, cmpars=cmpars, **kwa) # common mode correction
arrf = np.array(raw & cc.data_bit_mask(), dtype=np.float32) - pedest
print('== Event %04d ==' % self.counter)
# print('config', cc.config)
# print('calib_metadata', cc.calib_metadata('pedestals'))
print('calib_types', ctypes)
print(info_ndarr(peds, 'pedestals'))
print(info_ndarr(cc.gain(), 'gain'))
print(info_ndarr(gfactor, 'gain_factor'))
print(info_ndarr(status, 'status'))
print('common_mode from caliconst', str(comode))
print('number_of_panels', npanels)
print('trbit_for_panel(0)', trbit_p0)
print(info_ndarr(ascfg_p0, 'asicPixelConfig_for_panel(0)'))
print(info_ndarr(raw, 'raw'))
print(info_ndarr(mask, 'mask'))
print('dettype', dettype)
print(info_ndarr(cbitscfg, 'cbitscfg'))
print(info_ndarr(cbitstot, 'cbitstot'))
print(info_ndarr(gmaps, 'gmaps'))
print(info_ndarr(pedest, 'pedest'))
print(info_ndarr(factor, 'factor'))
print(info_ndarr(cmcorr, 'cmcorr'))
print(info_ndarr(arrf, 'raw(data bits)-peds'))
print(info_ndarr(calib0, 'calib0'))
# det.raw.calib(...) algorithm close reproduction
calib1 = None
if True:
arrf1 = arrf.copy()
cc.common_mode_apply(arrf1, gmaps, cmpars=cmpars, **kwa)
calib1 = arrf1 * factor if mask is None else arrf1 * factor * mask
print(info_ndarr(calib1, 'calib1'))
# det.raw.calib(...) - effective algorithm
calib2 = None
if True:
arrf2 = arrf.copy() + cmcorr
calib2 = arrf2 * factor if mask is None else arrf2 * factor * mask
print(info_ndarr(calib2, 'calib2'))
# img = cmcorr[0, 144:, :192]
# img = table_nxn_epix10ka_from_ndarr(cmcorr)
img = table_nxn_epix10ka_from_ndarr(calib1)
print(info_ndarr(img, 'img'))
return img |
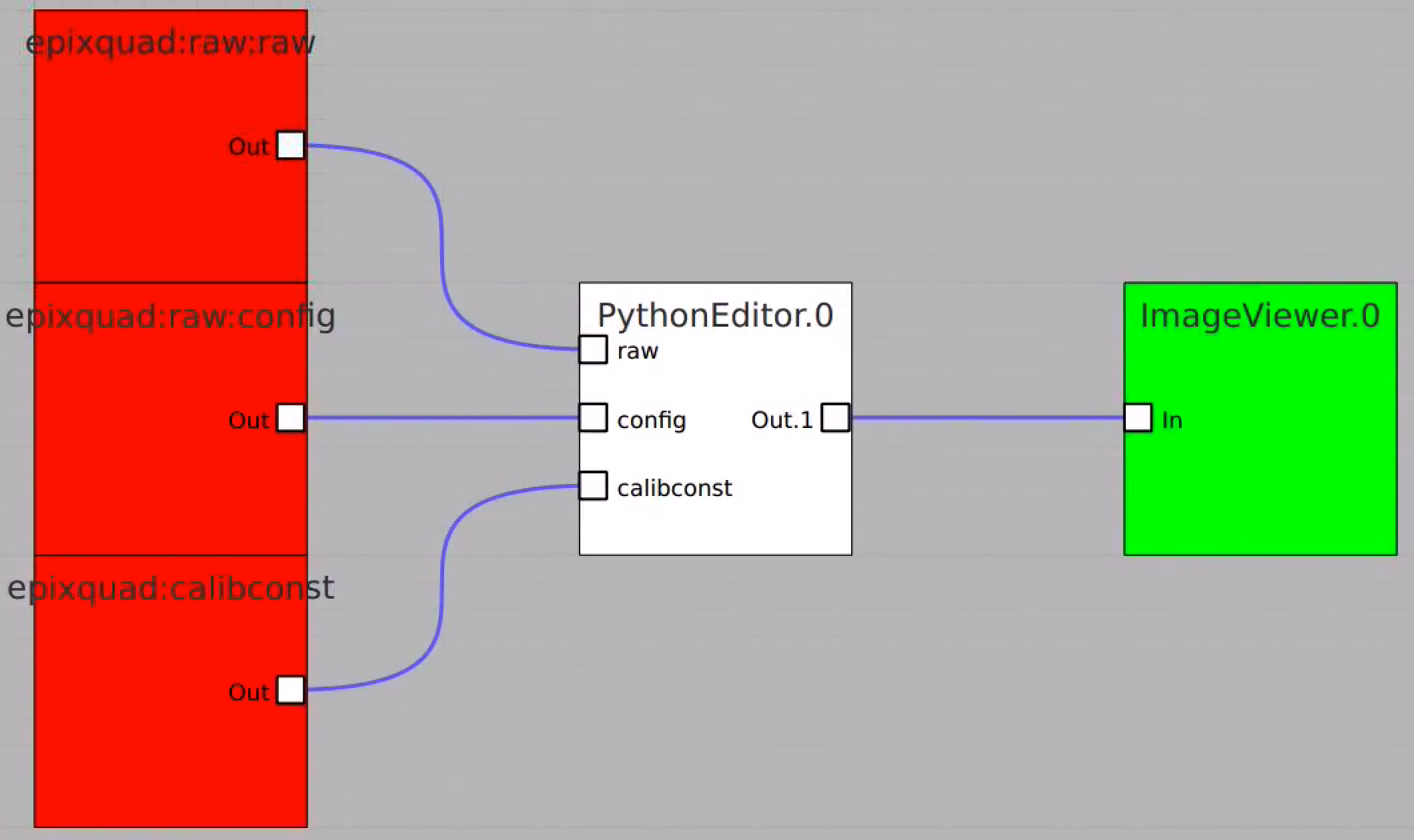
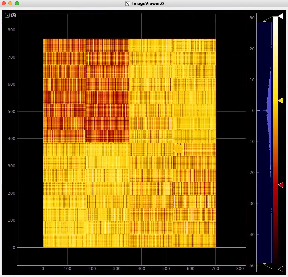
Image for common mode correction and calib1, respectively
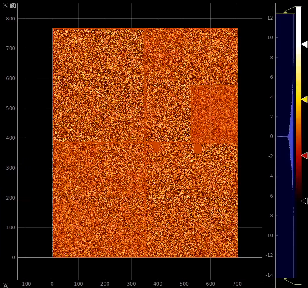
Example shows how to initialize and use the calib_components_epix object using parameters retrieved from the detector interface.
This script can be executed in the lcls2 psana environment by the command like
python ./lcls2/psana/psana/detector/test_issues_2023.py 2