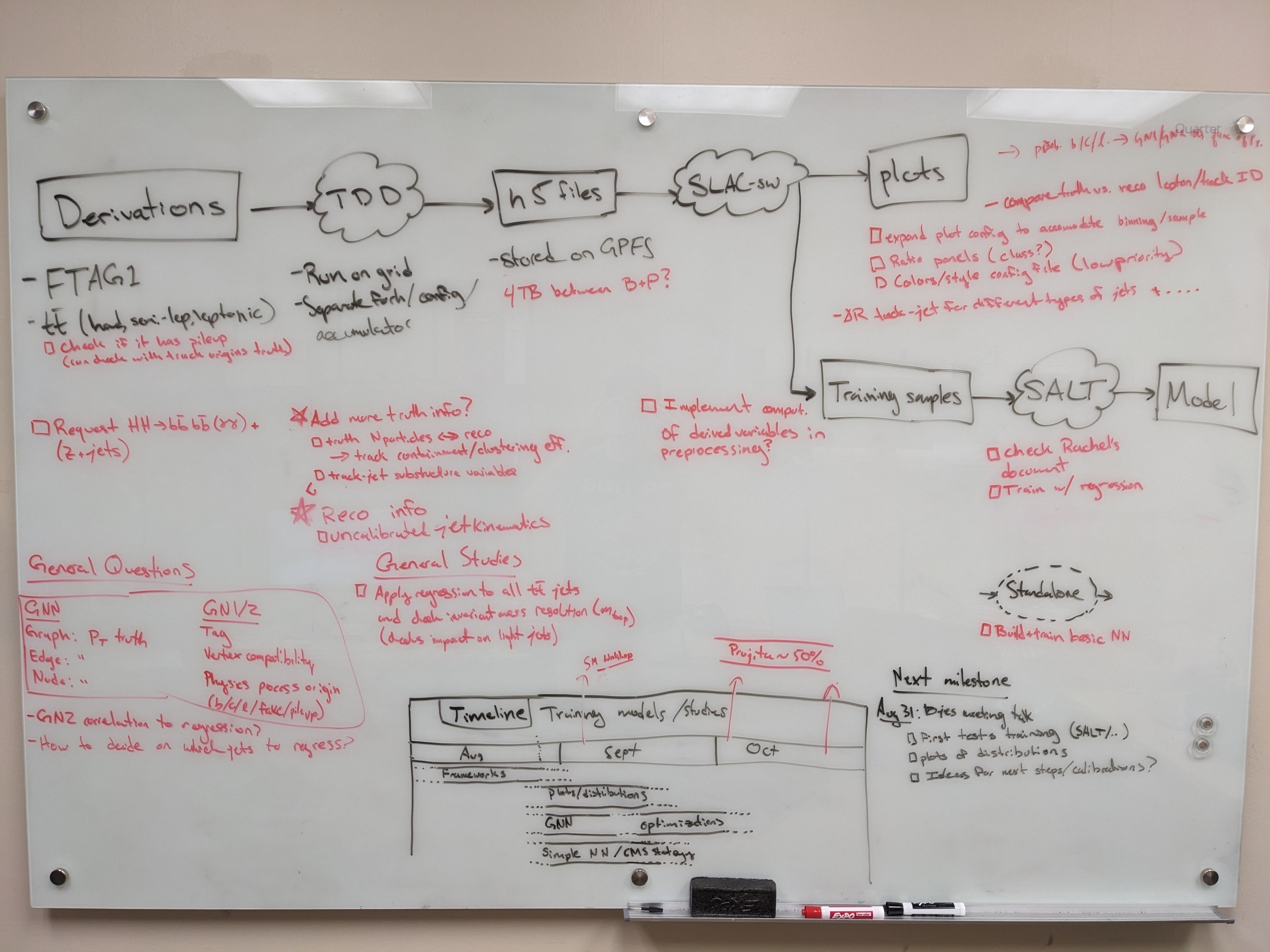
...
Planning
SDF preliminaries
Compute resources
SDF has a shared queue for submitting jobs via slurm, but this partition has extremely low priority. Instead, use the usatlas partition or request Michael Kagan to join the atlas partition.
You can use the command sacctmgr list qos name=atlas,usatlas format=GrpTRES%125 to check the resources available for each:
atlas has 2 CPU nodes (2x 128 cores) and 1 GPU node (4x A100); 3 TB memoryusatlas has 4 CPU nodes (4x 128 cores) and 1 GPU node (4x A100, 5x GTX 1080. Ti, 20x RTX 2080 Ti); 1.5 TB memory
Environments
An environment needs to be created to ensure all packages are available. We have explored some options for doing this.
Option 1: stealing instance built for SSI 2023. This installs most useful packages but uses python 3.6, which leads to issues with h5py.
Starting Jupyter sessions via SDF web interface
- SDF web interface > My Interactive Sessions > Services > Jupyter (starts a server via SLURM)
- Jupyter Instance > slac-ml/SSAI
Option 2 (preferred): create your own conda environment. Follow the SDF docs to use ATLAS group installation of conda.
There is also a slight hiccup with permissions in the folder /sdf/group/atlas/sw/conda/pkgs, which one can sidestep by specifying their own folder for saving packages (in GPFS data space).
The TLDR is:
...
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples ├── Rel22_ttbar_AllHadronic ├── Rel22_ttbar_DiLep └── Rel22_ttbar_SingleLep |
Preprocessing and plotting with Umami
SALT likes to take preprocesses data file formats from Umami. The best (read: only) way to use umami out of the box is via a docker container. To configure on SDF following the docs, add the following to your .bashrc:
| Code Block |
|---|
export SDIR=/scratch/${USER}/.singularity
mkdir $SDIR -p
export SINGULARITY_LOCALCACHEDIR=$SDIR
export SINGULARITY_CACHEDIR=$SDIR
export SINGULARITY_TMPDIR=$SDIR |
Since cvmfs is mounted on SDF, use the latest tagged version singularity container:
| Code Block |
|---|
singularity shell /cvmfs/unpacked.cern.ch/gitlab-registry.cern.ch/atlas-flavor-tagging-tools/algorithms/umami/umamibase-plus:0-20 |
There is no need to clone the umami repo unless you want to edit the code.
Preprocessing
An alternative (and supposedly better way to go) is to use the standalone umami-preprocessing (UPP).
Follow the installation instructions on git. This is unfortunately hosted on GitHub, so we cannot add it to the bjr-slac GitLab group
You can cut to the chase by doing conda activate upp.
Notes on logic of the preprocessing config files
There are several aspects of the preprocessing config that are not documented (yet). Most things are self explanatory, but note that within the FTag software,
there exist flavor classifications (for example lquarkjets, bjets, cjets, taujets, etc). These can be used to define different sample components. Further selections
based on kinematic cuts can be made through the region key. It appears only 2 features can be specified for the resampling (nominally pt and eta). The binning
also appears not to be respected.
Running SALT
The slac-bjr git project contains a fork of SALT. One can follow the SALT documentation for general installation/usage. Some specific notes can be found below:
Creating conda environment: One can use the environment salt that has been setup in /gpfs/slac/atlas/fs1/d/bbullard/conda_envs,
otherwise you may need to use conda install -c conda-forge jsonnet h5utils. Note that this was built using the latest master on Aug 28, 2023.
SALT also uses comet for logging, which has been installed into the salt environment (conda install -c anaconda -c conda-forge -c comet_ml comet_ml).
Follow additional instructions for configuration on the SALT documentation. Note that the following error may appear if following the normal installation:
| Code Block |
|---|
ImportError: cannot import name 'COMMON_SAFE_ASCII_CHARACTERS' from 'charset_normalizer.constant' |
To solve it, you can do the following (again, this is already available in the common salt environment):
| Code Block |
|---|
conda install chardet
conda install --force-reinstall -c conda-forge charset-normalizer=3.2.0 |
Testing SALT: it is ideal not to use interactive nodes to do training, but if this is needed for some validation you can use salt fit --config configs/GNX-bjr.yaml --trainer.fast_dev_run 2. If you get to the point where you see an
error that memory cannot be allocated, you at least vetted against other errors.
Analyzing H5 samples
Notebooks
...