...
The result is shown below. The first (on the left) image represents 32 so called 2x1 CSpad elements stacked into a 2D array. The second images represents a geometrically correct (including relative alignment of the elements) frame:
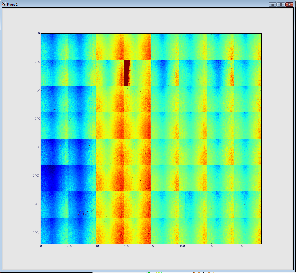
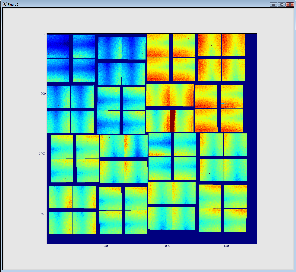
Advanced techniques
| Anchor |
|---|
| Advanced Techniques |
|---|
| Advanced Techniques |
|---|
|
Random Access to XTC Files ("Indexing")
LCLS runs which have XTC index files (generated by the DAQ) can have their events accessed in non-sequential order. The associated EPICS variables and beginCalib information is correctly provided by psana. You can check for the existence of these files by substituting your experiment name in a command similar to:
| Code Block |
|---|
|
ls /reg/d/psdm/XCS/xcs84213/xtc/index |
There should be 1 index file per data file in your experiment. If they do not exist, they can be created using an "xtcindex" command (send email to "pcds-help@slac.stanford.edu" to have this done).
| Code Block |
|---|
|
import psana
# note the "idx" input source at the end of this line: indicates that events will be accessed randomly.
ds = psana.DataSource('exp=cxib7913:run=34:idx')
for run in ds.runs():
# get array of timestamps of events
times = run.times()
for i in range(3,-1,-1):
#request a particular timestamp in a run
evt=run.event(times[i])
if evt is None:
print '*** event fetch failed'
continue
id = evt.get(psana.EventId)
print 'Fiducials:',id.fiducials() |
Some notes about indexing:
- If you have a list of timestamps for some other analysis, those can be used in the run.event() method without using the run.times() method.
- Index files are only made available by the DAQ at the end of the run, so can be used before then (this makes "realtime" FFB/shared-memory analysis impossible.
- Indexing only currently works with XTC files (no HDF5 support)
- Currently, indexing only has information about one run at a time (one cannot easily jump between events in different runs).
MPI Parallelization
Using the above indexing feature, it is possible to use MPI to have psana analyze events in parallel (this is useful for many, but not all, algorithms) by having different cores access different events. In principle this can work on thousands of cores. This requires the python package mpi4py (part of the psana release) and a compatible version of openmpi (not currently in the psana release). This is some sample code that sums images in a run:
| Code Block |
|---|
|
import psana
import numpy as np
import sys
from mpi4py import MPI
comm = MPI.COMM_WORLD
rank = comm.Get_rank()
size = comm.Get_size()
ds = psana.DataSource('exp=cxib7913:run=34:idx')
src = psana.Source('DetInfo(CxiDg2.0:Tm6740.0)')
for run in ds.runs():
times = run.times()
mylength = len(times)/size
mytimes= times[rank*mylength:(rank+1)*mylength]
for i in range(mylength):
evt = run.event(mytimes[i])
if evt is None:
print '*** event fetch failed'
continue
pulnix = evt.get(psana.Camera.FrameV1,src)
if pulnix is None:
print '*** failed to get pulnix'
continue
if 'sum' in locals():
sum+=pulnix.data16()
else:
sum=pulnix.data16()
id = evt.get(psana.EventId)
print 'rank',rank,'analyzed event with fiducials',id.fiducials()
print 'image:\n',pulnix.data16()
sumall = np.empty_like(sum)
#sum the images across mpi cores
comm.Reduce(sum,sumall)
if rank==0:
print 'sum is:\n',sumall |
This can be interactively parallelizing over 2 cores with a command similar to the following:
| Code Block |
|---|
/reg/neh/home1/cpo/junk/openmpi-1.8/install/bin/mpirun -n 2 python mpi.py |
or run in a batch job:
| Code Block |
|---|
bsub -a mympi -n 2 -o mpi.log -q psfehmpiq python mpi.py |
Re-opening data sets, opening multiple data sets simultaneously
...

