...
- SDF guide and documentation, particularly on using Jupyter notebooks interactively or through web interface (runs on top of nodes managed by SLURM)
- Training dataset dumper (used for producing h5 files from FTAG derivations) documentation and git (Prajita's fork, bjet_regression is the main branch)
- SALT documentation, SALT on SDF, puma git repo (used for plotting), and Umami docs (for postprocessing), also umami-preprocessing (UPP)
- SLAC GitLab group for plotting related code
Code Block export PATH="/sdf/group/atlas/sw/conda/bin:$PATH"FTAG1 derivation definition (FTAG1.py)
Documents and notes
- GN1 June 2022 PUB note, nice slides and proceedings from A. Duperrin
- Jannicke's thesis (chapter 4 on b-jets)
...
- Understanding bias in JES in low truth-jet pT
- Truth-jet matching (Brendon)
- Check different truth jet matching schemes for response definition
- Performance as a function of dR quality cut
- Check different truth jet matching schemes for response definition
- Evaluating the impact of soft lepton information (Prajita)
- Performance with LeptonID
- Apply additional selection on soft electron
- Auxiliary tasks (Brendon)
- Fixing weights of TruthOrigin classes
- Add TruthParticleID
- Model architecture (Brendon)
- Attention mechanisms
- Hyperparameter optimization
...
atlas has 2 CPU nodes (2x 128 cores) and 1 GPU node (4x A100); 3 TB memoryusatlas has 4 CPU nodes (4x 128 cores) and 1 GPU node (4x A100, 5x GTX 1080. Ti, 20x RTX 2080 Ti); 1.5 TB memory
Environments
For sdf
An environment needs to be created to ensure all packages are available. We have explored some options for doing this.
Option 1: stealing instance built for SSI 2023. This installs most useful packages but uses python 3.6, which leads to issues with h5py.
Starting Jupyter sessions via SDF web interface
...
| Code Block |
|---|
export PATH="/sdf/group/atlas/sw/conda/bin:$PATH"
export TMPDIR="${SCRATCH}" |
Producing H5 samples with Training Dataset Dumper
Introduction
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Overview (FTag Workshop Sept. 2023): presentation by Nikita Pond
Tutorial: TDD on FTag docs
General introduction to Athena in Run 3 (component accumulator): ATLAS Software Docs
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
TDD productions:
v2.1: TDD code, BigPanda
Run with grid-submit -c configs/PHYSVAL_EMPFlow_jer.json -e AntiKt4EMPFlow_PtReco_Correction_MV2c10_DL1r_05032020.root -i BTagTrainingPreprocessing/grid/inputs/physval-jer.txt bjer
NOTE: you will need to copy the PtReco histogram into the top level of training-dataset-dumper
- Issue with datasets not being available on the grid.. need to request a new production
- GSC pre-recommendation (gitlab), jet calibrations in Athena
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
For S3DF
Setting up the conda environment has steps similar to those of SDF. To use the atlas group conda, use following steps
1.) Add the following in your .bashrc
| Code Block |
|---|
export PATH="/sdf/group/atlas/sw/conda/bin:$PATH" |
2.) Add the following to your .condarc
| Code Block |
|---|
pkgs_dirs:
- /fs/ddn/sdf/group/atlas/d/<user>/conda_envs/pkgs
envs_dirs:
- /fs/ddn/sdf/group/atlas/d/<user>/conda_envs/envs
channels:
- conda-forge
- defaults
- anaconda
- pytorch |
3.) source conda.sh
| Code Block |
|---|
> source /sdf/group/atlas/sw/conda/etc/profile.d/conda.sh |
4.) cross check if conda is set correctly
| Code Block |
|---|
> which conda
#following should appear if things went smoothly
conda ()
{
\local cmd="${1-__missing__}";
case "$cmd" in
activate | deactivate)
__conda_activate "$@"
;;
install | update | upgrade | remove | uninstall)
__conda_exe "$@" || \return;
__conda_reactivate
;;
*)
__conda_exe "$@"
;;
esac
} |
5.) Now follow the same steps as SDF to create and activate the conda environment
...
Producing H5 samples with Training Dataset Dumper
Introduction
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Overview (FTag Workshop Sept. 2023): presentation by Nikita Pond
Tutorial: TDD on FTag docs
General introduction to Athena in Run 3 (component accumulator): ATLAS Software Docs
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
TDD productions:
v2.1: TDD code, BigPanda
Run with grid-submit -c configs/PHYSVAL_EMPFlow_jer.json -e AntiKt4EMPFlow_PtReco_Correction_MV2c10_DL1r_05032020.root -i BTagTrainingPreprocessing/grid/inputs/physval-jer.txt bjer
NOTE: you will need to copy the PtReco histogram into the top level of training-dataset-dumper
- Issue with datasets not being available on the grid.. need to request a new production
- GSC pre-recommendation (gitlab), jet calibrations in Athena
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
We are using a custom fork of training-dataset-dumper, which is used to extract h5 files from derivations - we are currently using FTAG1 / PHYSVAL / JETM2.
The fork in slac-We are using a custom fork of training-dataset-dumper, which is used to extract h5 files from derivations - we are currently using FTAG1 / PHYSVAL / JETM2.
The fork in slac-bjr has our working code to dump datasets based on the AntiKt4TruthDressedWZJets container and EMPFlow as well as UFO jets.
...
| Code Block | ||
|---|---|---|
| ||
#SBATCH --output="<your_output_path>/out/slurm-%j.%x.out"
#SBATCH --error="<your_output_path>/out/slurm-%j.%x.err
export COMET_API_KEY=<your_commet_key>
export COMET_WORKSPACE=<your_commet_workspace>
export COMET_PROJECT_NAME=<your_project_name>
cd <your_path_to_salt_directory> |
from top level salt directory can use following command to launch a slurm training job in sdf
| Code Block | ||
|---|---|---|
| ||
sbatch salt/submit_slurm_bjr.sh |
Both slurm-%j.%x.err andslurm-%j.%x.out files will start to fill up
You can use standard sbatch commands from SDF documentation to understand the state of your job.
Comet Training Visualization
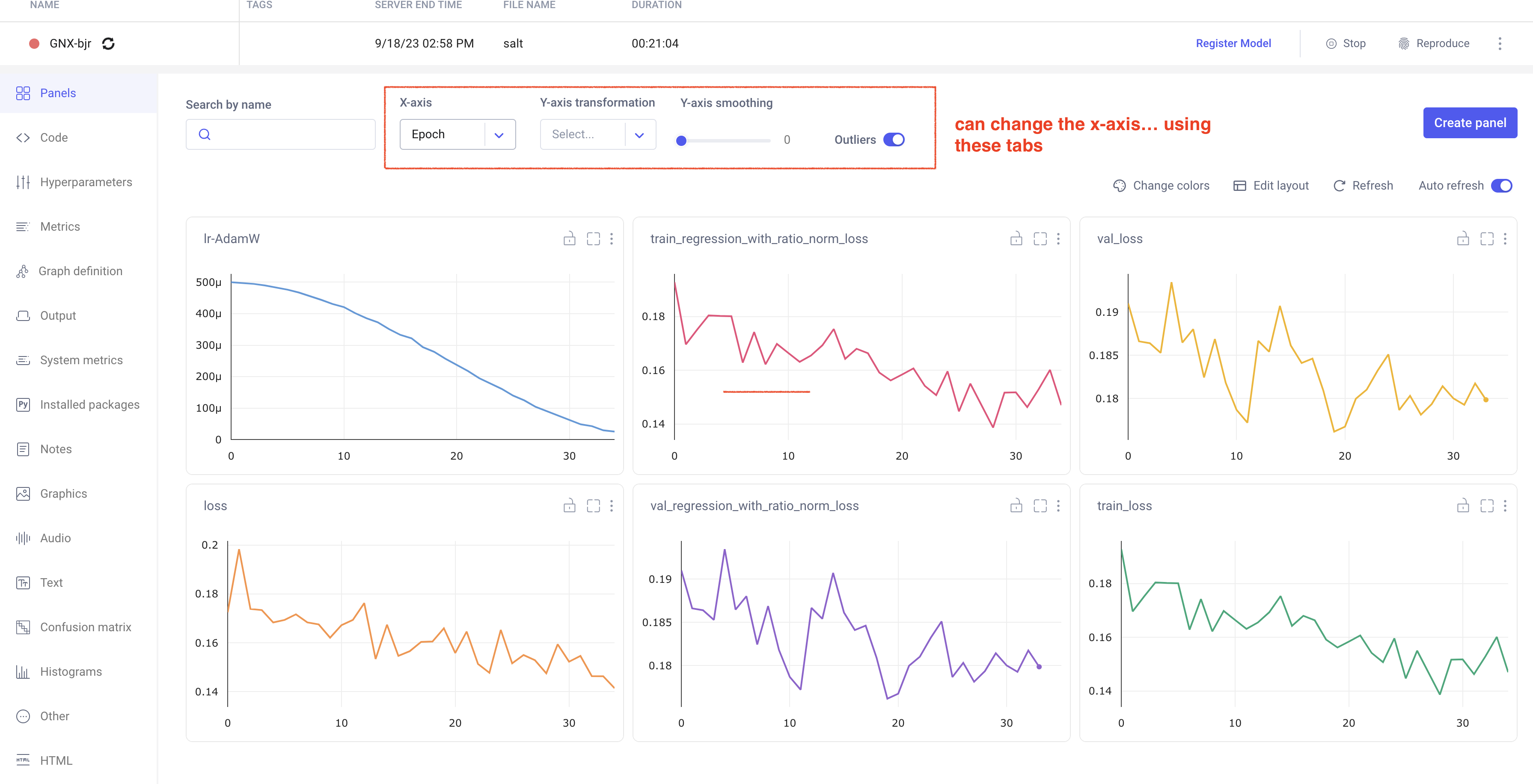
In your comet profile, you should start seeing the live update for the training which looks as follows. The project name you have specified in the submit script appears under your
workspace which you can click to get the graphs of live training updates.
Training Evaluation
Follow salt documentation to run the evaluation of the trained model in the test dataset. There is a separate batch submission script used for the model evaluation, but is very similar to what is used in the model training batch script.
The main difference is in the salt command that is run (see below). It will produce a log in the same directory as the other log files, and will produce a new output h5 file alongside the one you pass in for evaluation.
The important points are the following:
- --data.test_suff: Specify a suffix for the sample that is produced from the PredictionWriter callback specified in the model config. There is a separate list of features to be saved to this new file, along with the model output,
that can be used for studies of the model performance - --data.num_workers: you should use the same number of workers for the evaluation as for the training, since both are bottlenecked by the loading of the data
- --data.test_file: technically can be either the training, testing, or evaluation h5 file. In principle the testing file is created for this purpose. Philosophically, you want to keep this dataset separate so that you don't induce some kind of bias as you manually determine the hyper parameter optimization
- --ckpt_path: the specific model checkpoint you want to use. Out of the box, SALT should be picking the checkpoint with the lowest validation loss, but this has been found to not be very reliable. So always do it manually to be sure you know what model state you are actually studying.
| Code Block |
|---|
srun salt test -c logs/<model>/config.yaml --data.test_file <path_to_file_for_evaluation>.h5 --data.num_jets_train <num_jets> --data.num_workers 20 --trainer.devices 1 --data.test_suff eval --ckpt_path logs/<model>/ckpts/epoch=<epoch>.ckpt |
We are developing some baseline evaluation classes in the analysis repository to systematically evaluate model performance.
To do: develop this, and add some notes on it (Brendon/Prajita)
Miscellaneous tips
You can grant read/write access for GPFS data folder directories to ATLAS group members via the following (note that this does not work for SDF home folder)
=<your_commet_workspace>
export COMET_PROJECT_NAME=<your_project_name>
cd <your_path_to_salt_directory> |
from top level salt directory can use following command to launch a slurm training job in sdf
| Code Block | ||
|---|---|---|
| ||
sbatch salt/submit_slurm_bjr.sh |
Both slurm-%j.%x.err andslurm-%j.%x.out files will start to fill up
You can use standard sbatch commands from SDF documentation to understand the state of your job.
Comet Training Visualization
In your comet profile, you should start seeing the live update for the training which looks as follows. The project name you have specified in the submit script appears under your
workspace which you can click to get the graphs of live training updates.
Training Evaluation
Follow salt documentation to run the evaluation of the trained model in the test dataset. There is a separate batch submission script used for the model evaluation, but is very similar to what is used in the model training batch script.
The main difference is in the salt command that is run (see below). It will produce a log in the same directory as the other log files, and will produce a new output h5 file alongside the one you pass in for evaluation.
The important points are the following:
- --data.test_suff: Specify a suffix for the sample that is produced from the PredictionWriter callback specified in the model config. There is a separate list of features to be saved to this new file, along with the model output,
that can be used for studies of the model performance - --data.num_workers: you should use the same number of workers for the evaluation as for the training, since both are bottlenecked by the loading of the data
- --data.test_file: technically can be either the training, testing, or evaluation h5 file. In principle the testing file is created for this purpose. Philosophically, you want to keep this dataset separate so that you don't induce some kind of bias as you manually determine the hyper parameter optimization
- --ckpt_path: the specific model checkpoint you want to use. Out of the box, SALT should be picking the checkpoint with the lowest validation loss, but this has been found to not be very reliable. So always do it manually to be sure you know what model state you are actually studying.
| Code Block |
|---|
srun salt test -c logs/<model>/config.yaml --data.test_file <path_to_file_for_evaluation>.h5 --data.num_jets_train <num_jets> --data.num_workers 20 --trainer.devices 1 --data.test_suff eval --ckpt_path logs/<model>/ckpts/epoch=<epoch>.ckpt |
We are developing some baseline evaluation classes in the analysis repository to systematically evaluate model performance.
To do: develop this, and add some notes on it (Brendon/Prajita)
...
Miscellaneous tips
You can grant read/write access for GPFS data folder directories to ATLAS group members via the following (note that this does not work for SDF home folder)
| Code Block |
|---|
groups <username> # To check user groups
cd <your_directory>
find . -type d|xargs chmod g+rx # Need to make all subdirectories readable *and* executable to the group |
You can setup passwordless ssh connection following this page to create an ssh key pair. Then add this block to your ~/.ssh/config file (allows to tunnel directly to iana, but you can adapt to other hosts):
| Code Block |
|---|
Host sdfiana
HostName iana
User <yourusername>
IdentityFile ~/.ssh/<your private key>
proxycommand ssh s3dflogin.slac.stanford.edu nc %h %p
ForwardX11 yes
Host s3dflogin.slac.stanford.edu |
| Code Block |
groups <username> # To check user groups
cd <your_directory>
find . -type d|xargs chmod g+rx # Need to make all subdirectories readable *and* executable to the group |
Discussion Summary
Suggestions from M. Kagan Jan 23, 2023
...