...
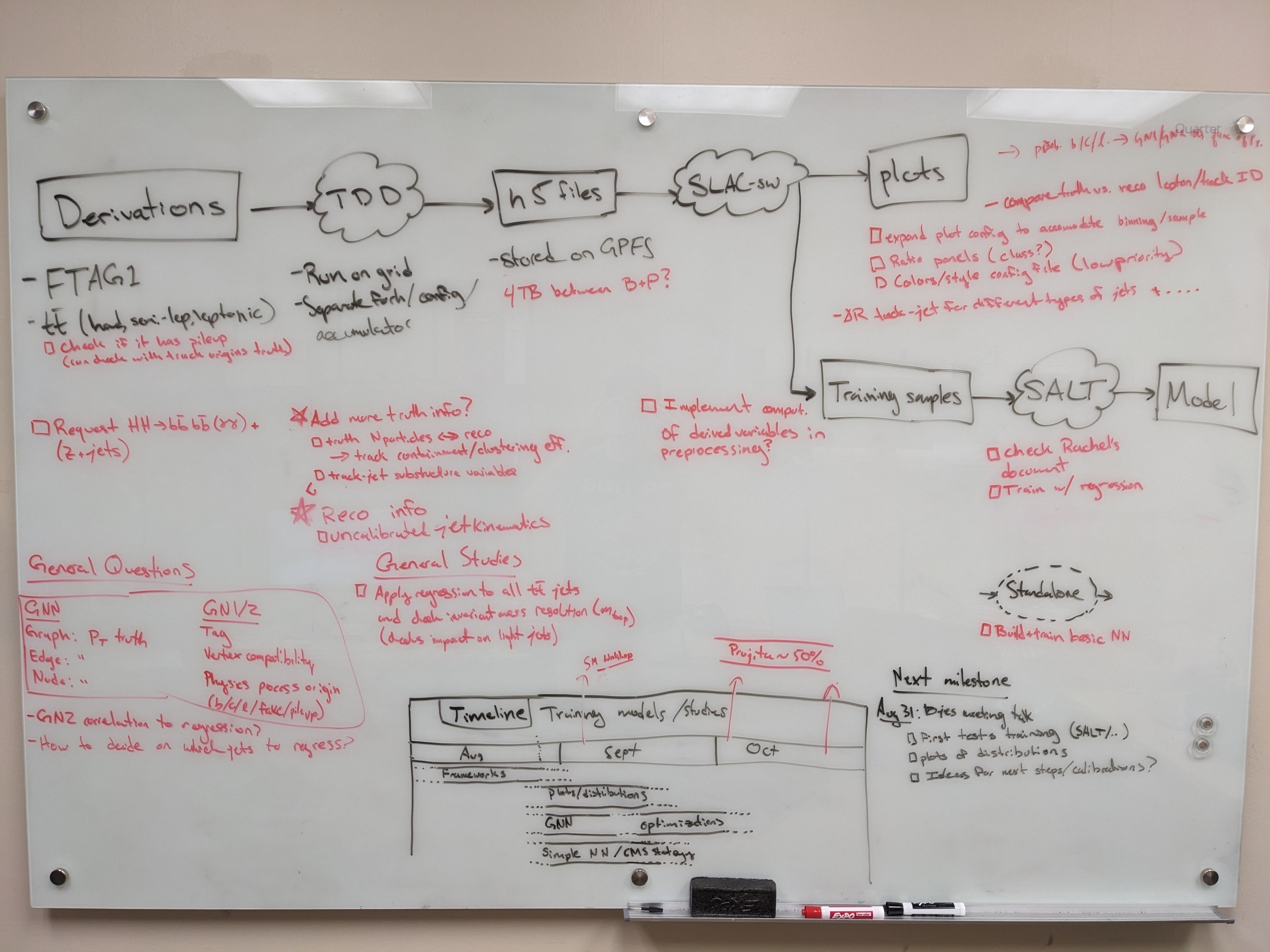
Planning
SDF preliminaries
Compute resources
SDF has a shared queue for submitting jobs via slurm, but this partition has extremely low priority. Instead, use the usatlas partition or request Michael Kagan to join the atlas partition.
...
atlas has 2 CPU nodes (2x 128 cores) and 1 GPU node (4x A100); 3 TB memoryusatlas has 4 CPU nodes (4x 128 cores) and 1 GPU node (4x A100, 5x GTX 1080. Ti, 20x RTX 2080 Ti); 1.5 TB memory
Environments
An environment needs to be created to ensure all packages are available. We have explored some options for doing this.
Option 1: stealing instance built for SSI 2023. This installs most useful packages but uses python 3.6, which leads to issues with h5py.
Starting Jupyter sessions via SDF web interface
...
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/bbullard/conda_env_yaml/ ├── puma.yaml ├── salt.yaml └── upp.yaml |
Producing H5 samples with Training Dataset Dumper
We are using a custom fork of training-dataset-dumper, developed for producing h5 files for NN training based on FTAG derivations.
The custom fork is modified to store the truth jet pT via AntiKt4TruthDressedWZJets container.
...
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples ├── nonallhad_PHYSVAL_mc20_410470 # High stats PHYSVAL derivation ├── Rel22_ttbar_AllHadronic ├── Rel22_ttbar_DiLep └── Rel22_ttbar_SingleLep |
To do: need to add a lot of details on this (Prajita)
- Links to specific component accumulators and CP tools needed to get (soft) muon/electron information
- Details on calibrations used for variables in the training
Plotting with Umami/Puma (Work In Progress)
NOTE: we have not yet used umami/puma for plotting or preprocessing, but it can in principle be done and we will probably try to get this going soon.
Plotting with umami
Umami (which relies on puma internally) is capable of producing plots based on yaml configuration files.
The best (read: only) way to use umami out of the box is via a docker container. To configure on SDF following the docs, add the following to your .bashrc:
...
Plotting exercises can be followed in the umami tutorial.
Plotting with puma standalone
Puma can be used to produce plots in a more manual way. To install, there was some difficulty following the nominal instructions in the docs. What I (Brendon) found to work was to do:
...
The slac-bjr git project contains a fork of SALT. One can follow the SALT documentation for general installation/usage. Some specific notes can be found below:
Creating conda environment
One can use the environment salt that has been setup in /gpfs/slac/atlas/fs1/d/bbullard/conda_envs,
otherwise you may need to use conda install -c conda-forge jsonnet h5utils. Note that this was built using the latest master on Aug 28, 2023.
...
| Code Block |
|---|
conda install chardet conda install --force-reinstall -c conda-forge charset-normalizer=3.2.0 |
Interactive testing
It is suggested not to use interactive nodes to do training, but instead to open a terminal on an SDF node by:
...
| Code Block |
|---|
salt fit -c configs/<my_config>.yaml --data.num_jets_train <small_number> --data.num_workers <num_workers> |
For training on slurm
See the SALT on SDF documentation (also linked at the top of this page) and example configs in the SALT fork in the slac_bjr GitLab project.
For additional clarity, see the following description of the submission scripts. Change the submit_slurm.sh script as follows
...
You can use standard sbatch commands from SDF documentation to understand the state of your job.
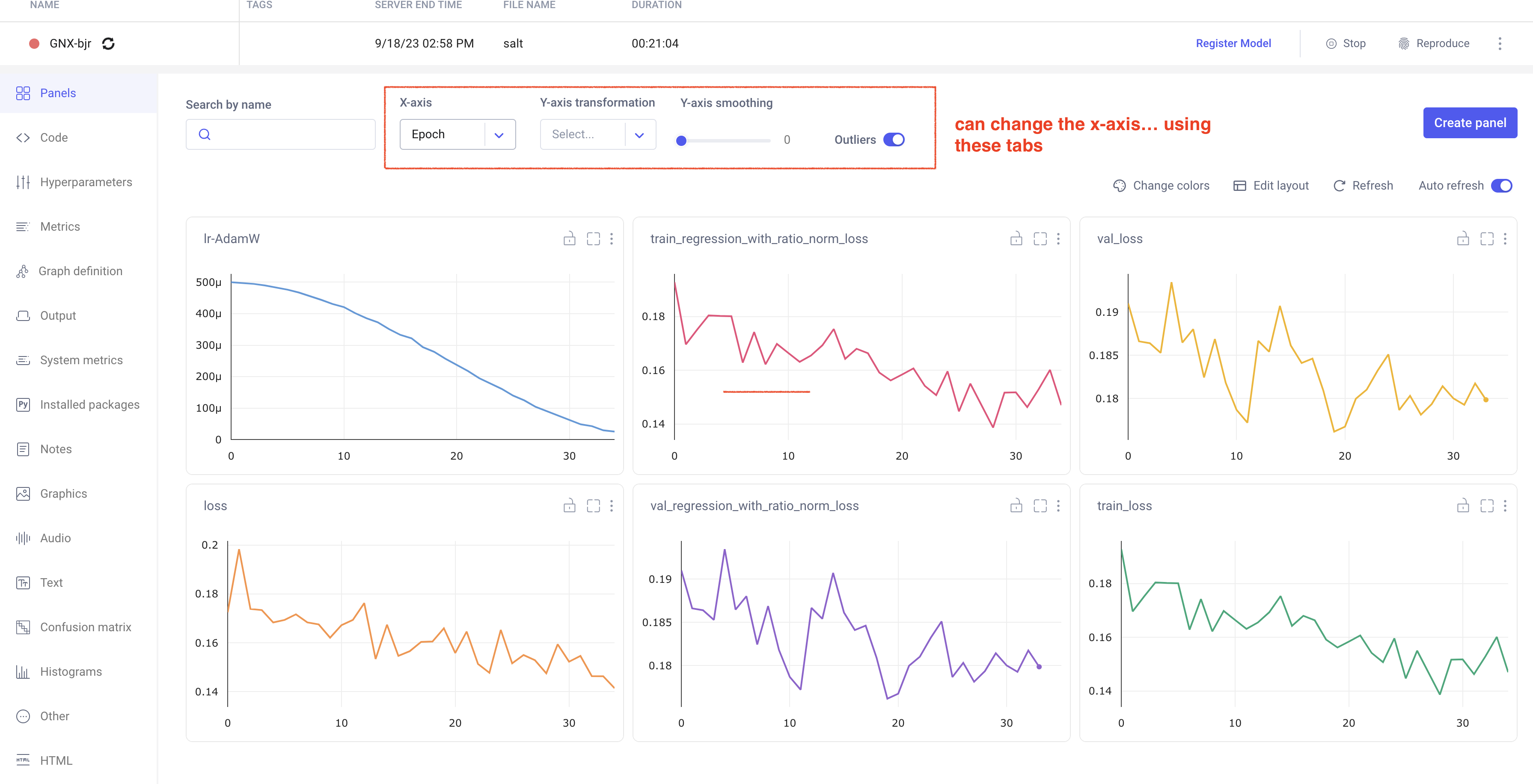
Comet Training Visualization
In your comet profile, you should start seeing the live update for the training which looks as follows. The project name you have specified in the submit script appears under your
workspace which you can click to get the graphs of live training updates.
Training Evaluation
Follow salt documentation to run the evaluation of the trained model in the test dataset. There is a separate batch submission script used for the model evaluation, but is very similar to what is used in the model training batch script.
The main difference is in the salt command that is run (see below). It will produce a log in the same directory as the other log files, and will produce a new output h5 file alongside the one you pass in for evaluation.
...
We are developing some baseline evaluation classes in the analysis repository to systematically evaluate model performance.
To do: add a link to the
...
develop this, and add some notes on it (Brendon/Prajita)
Miscellaneous tips
You can grant read/write access for GPFS data folder directories to ATLAS group members via the following (note that this does not work for SDF home folder)
...