...
| Code Block |
|---|
export CONDA_PREFIX=/sdf/group/atlas/sw/conda
export PATH=${CONDA_PREFIX}/bin/:$PATH
source ${CONDA_PREFIX}/etc/profile.d/conda.sh
conda env list
conda activate bjr_v01 |
For the following packages (UPP, SALT, and puma) there are predefined conda environments available (made by Brendon). You can activate them with the usual conda activate command. They are here:
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/bbullard/conda_env_yaml/
├── puma.yaml
├── salt.yaml
└── upp.yaml |
Producing H5 samples
We are using a custom fork of training-dataset-dumper, developed for producing h5 files for NN training based on FTAG derivations.
The custom fork is modified to store the truth jet pT via AntiKt4TruthDressedWZJets container.
...
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples ├── Rel22_ttbar_AllHadronic ├── Rel22_ttbar_DiLep └── Rel22_ttbar_SingleLep |
Plotting with
...
Umami/Puma (Work In Progress)
NOTE: we have not yet used umami/puma for plotting or preprocessing, but it can in principle be done and we will probably try to get this going soon.
Plotting with umami
Umami (which relies on puma internally) is capable of producing plots based on yaml configuration files.
The best (read: only) way to use umami out of the box is via a docker container. To configure on SDF following the docs, add the following to your .bashrc:
...
Interactive testing: it is suggested not to use interactive nodes to do training, but instead to open a terminal on an SDF node by starting a Jupyter notebook and going to New > Terminal. From here, you can test your training configuration.
Be aware that running a terminal on an SDF node will reduce your priority when submitting jobs to slurm. You also need to choose a conda environment that has jupyter installed, and then you can create a terminal and start whatever conda env you want!
Note that you must be careful about the number of workers you select (in the PyTorch trainer object) which should be <= the number of CPU cores you're using (using more CPU cores parallelizes the data loading,
which can be the primary bottleneck in training). The number of requested GPUs should match the number of devices used in the training.
...
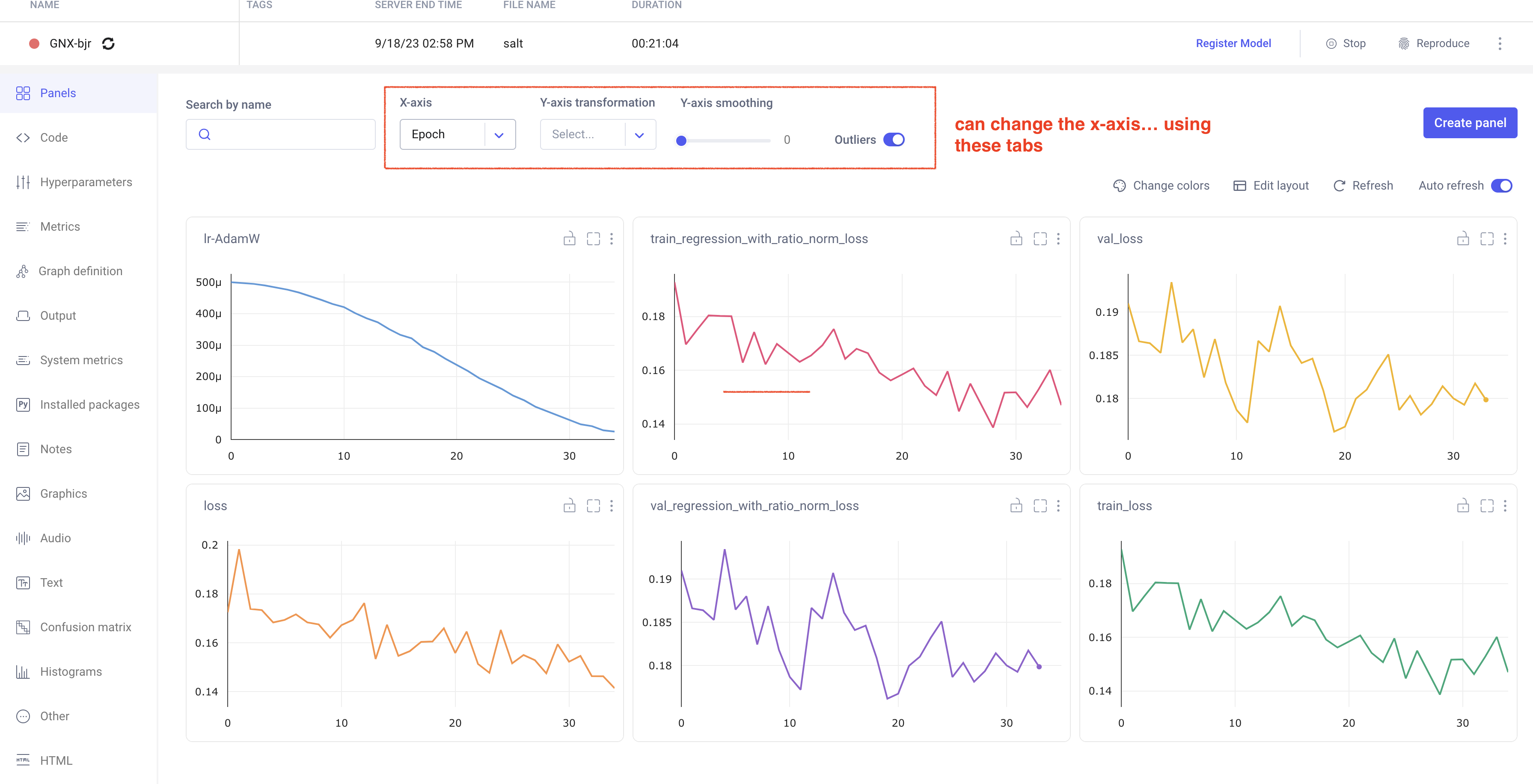
In your comet profile, you should start seeing the live update for the training which looks as follows. The project name you have specified in the submit script appears under your workspace which you can click to get the graphs of live training updates.
Training Evaluation
Follow salt documentation to run the evaluation of the trained model in the test dataset. This can also be done using a batch system. I (Prajita) personally have a separate bash script similar to the training script, for evaluating the training. The script looks like the following.
...