Table of Contents
| Table of Contents |
|---|
Useful links
Technical
- SDF guide and documentation, particularly on using Jupyter notebooks interactively or through web interface (runs on top of nodes managed by SLURM)
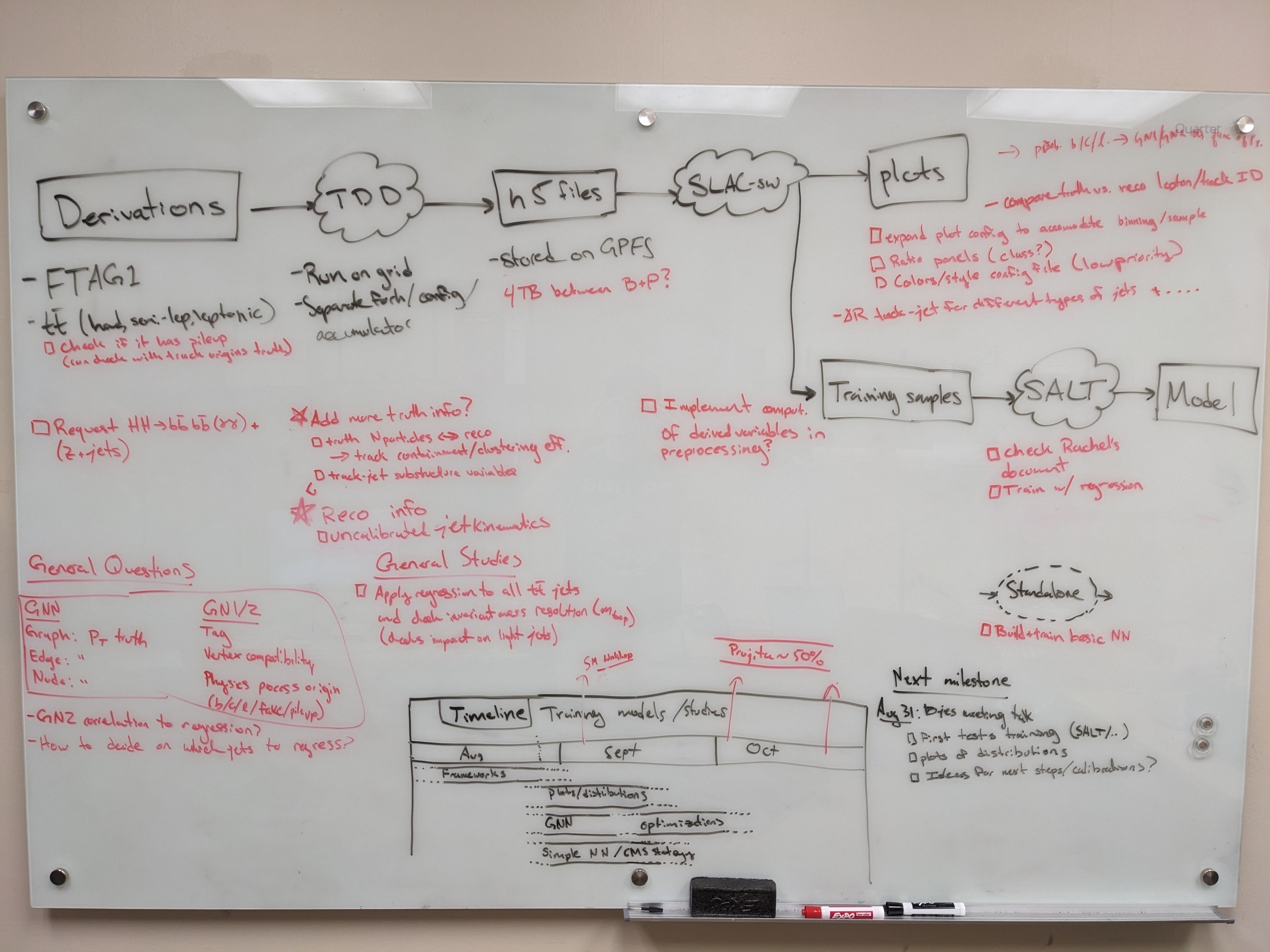
- Training dataset dumper (used for producing h5 files from FTAG derivations) documentation and git (Prajita's fork, bjet_regression is the main branch)
- SALT documentation, SALT on SDF, puma git repo (used for plotting), and Umami docs (for postprocessing), also umami-preprocessing (UPP)
- SLAC GitLab group for plotting related code
- FTAG1 derivation definition (FTAG1.py)
...
- See all B-jet calibration meetings on Indico
- Framework experience (Prajita, July 6)
- Plans (Prajita, July 13)
- What needs to be added to JETM2 (August 17)
Planning
SDF preliminaries
Compute resources
SDF has a shared queue for submitting jobs via slurm, but this partition has extremely low priority. Instead, use the usatlas partition or request Michael Kagan to join the atlas partition.
...
| Code Block |
|---|
export CONDA_PREFIX=/sdf/group/atlas/sw/conda
export PATH=${CONDA_PREFIX}/bin/:$PATH
source ${CONDA_PREFIX}/etc/profile.d/conda.sh
conda env list
conda activate bjr_v01 |
Producing H5 samples
We are using a custom fork of training-dataset-dumper, developed for producing h5 files for NN training based on FTAG derivations.
The custom fork is modified to store the truth jet pT via AntiKt4TruthDressedWZJets container.
...
| Code Block |
|---|
/gpfs/slac/atlas/fs1/d/pbhattar/BjetRegression/Input_Ftag_Ntuples ├── Rel22_ttbar_AllHadronic ├── Rel22_ttbar_DiLep └── Rel22_ttbar_SingleLep |
Plotting with Umami/Puma
Plotting with umami
Umami (which relies on puma internally) is capable of producing plots based on yaml configuration files.
The best (read: only) way to use umami out of the box is via a docker container. To configure on SDF following the docs, add the following to your .bashrc:
...
This took quite some time to run, so (again) save yourself the effort and use the precompiled environments.
Preprocessing
SALT likes to take preprocesses data file formats from Umami (though in principle the format is the same as what's produced by the training dataset dumper).
...
- within the FTag software, there exist flavor classifications (for example lquarkjets, bjets, cjets, taujets, etc). These can be used to define different sample components. Further selections
based on kinematic cuts can be made through theregionkey. - It appears only 2 features can be specified for the resampling (nominally pt and eta).
- The binning also appears not to be respected for the variables used in resampling.
Model development with SALT
The slac-bjr git project contains a fork of SALT. One can follow the SALT documentation for general installation/usage. Some specific notes can be found below:
...
Note that you must be careful about the number of workers you select (in the PyTorch trainer object) which should be <= the number of CPU cores you're using (using more CPU cores parallelizes the data loading,
which can be the primary bottleneck in training). The number of requested GPUs should match the number of devices used in the training.
Miscellaneous tips
You can grant read/write access for GPFS data folder directories to ATLAS group members via the following (note that this does not work for SDF home folder)
...